Gene Page: ITPKB
Summary ?
| GeneID | 3707 |
| Symbol | ITPKB |
| Synonyms | IP3-3KB|IP3K|IP3K-B|IP3KB|PIG37 |
| Description | inositol-trisphosphate 3-kinase B |
| Reference | MIM:147522|HGNC:HGNC:6179|Ensembl:ENSG00000143772|HPRD:00942|Vega:OTTHUMG00000037582 |
| Gene type | protein-coding |
| Map location | 1q42.13 |
| Sherlock p-value | 0.576 |
| Fetal beta | -1.905 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0217 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02814998 | 1 | 226926791 | ITPKB | 5.539E-4 | -0.374 | 0.049 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4501739 | chrX | 35960848 | ITPKB | 3707 | 0.01 | trans |
Section II. Transcriptome annotation
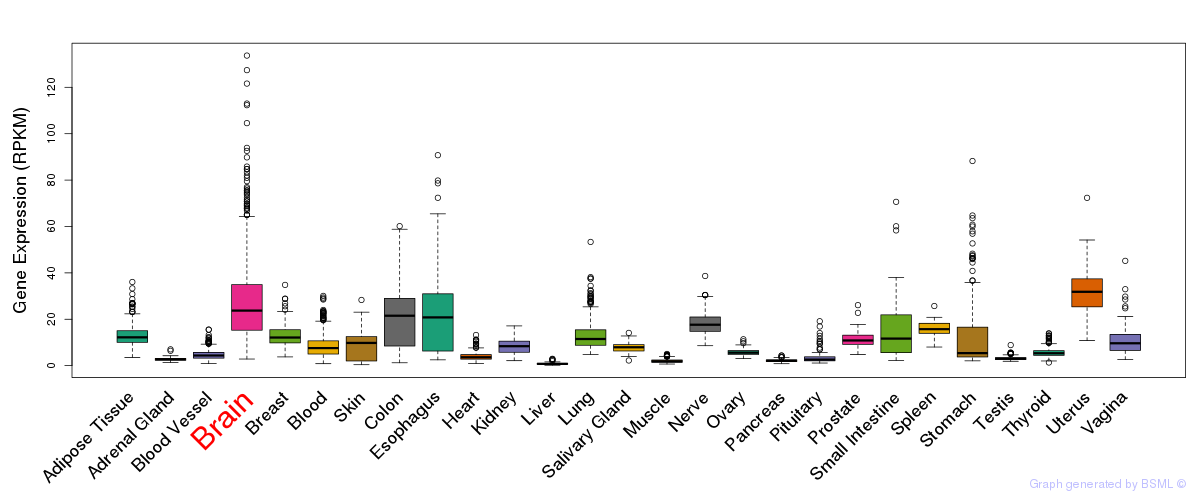
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHST6 | 0.78 | 0.79 |
| ANGPTL2 | 0.78 | 0.82 |
| OLIG2 | 0.77 | 0.81 |
| DOCK1 | 0.76 | 0.78 |
| ITPKB | 0.76 | 0.81 |
| SLC12A4 | 0.76 | 0.77 |
| LITAF | 0.75 | 0.77 |
| MID1IP1 | 0.75 | 0.78 |
| SOX13 | 0.75 | 0.81 |
| ST5 | 0.75 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPR22 | -0.47 | -0.51 |
| KLHL1 | -0.41 | -0.39 |
| NEUROD6 | -0.41 | -0.45 |
| MPPED1 | -0.41 | -0.43 |
| DACT1 | -0.41 | -0.35 |
| NELL2 | -0.40 | -0.38 |
| TIAM2 | -0.40 | -0.38 |
| SSBP2 | -0.40 | -0.39 |
| PPAPR5 | -0.40 | -0.40 |
| NR2C2AP | -0.40 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008440 | inositol trisphosphate 3-kinase activity | TAS | 1654894 | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 1654894 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| ST WNT CA2 CYCLIC GMP PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST GAQ PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ST PAC1 RECEPTOR PATHWAY | 7 | 5 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 109 | 115 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-132/212 | 2912 | 2918 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-142-3p | 111 | 117 | 1A | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-19 | 2588 | 2594 | 1A | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-194 | 2913 | 2920 | 1A,m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-375 | 1943 | 1949 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-410 | 2560 | 2566 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.