Gene Page: ITPR1
Summary ?
| GeneID | 3708 |
| Symbol | ITPR1 |
| Synonyms | ACV|CLA4|INSP3R1|IP3R|IP3R1|PPP1R94|SCA15|SCA16|SCA29 |
| Description | inositol 1,4,5-trisphosphate receptor type 1 |
| Reference | MIM:147265|HGNC:HGNC:6180|Ensembl:ENSG00000150995|HPRD:00925|Vega:OTTHUMG00000154996 |
| Gene type | protein-coding |
| Map location | 3p26.1 |
| Pascal p-value | 0.081 |
| Sherlock p-value | 0.669 |
| TADA p-value | 0.032 |
| Fetal beta | -1.681 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_mGluR5 G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITPR1 | chr3 | 4715041 | C | T | NM_001099952 NM_001168272 NM_002222 | p.809S>L p.794S>L p.794S>L | missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26379672 | 3 | 4534954 | ITPR1 | 4.96E-10 | -0.013 | 8.58E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
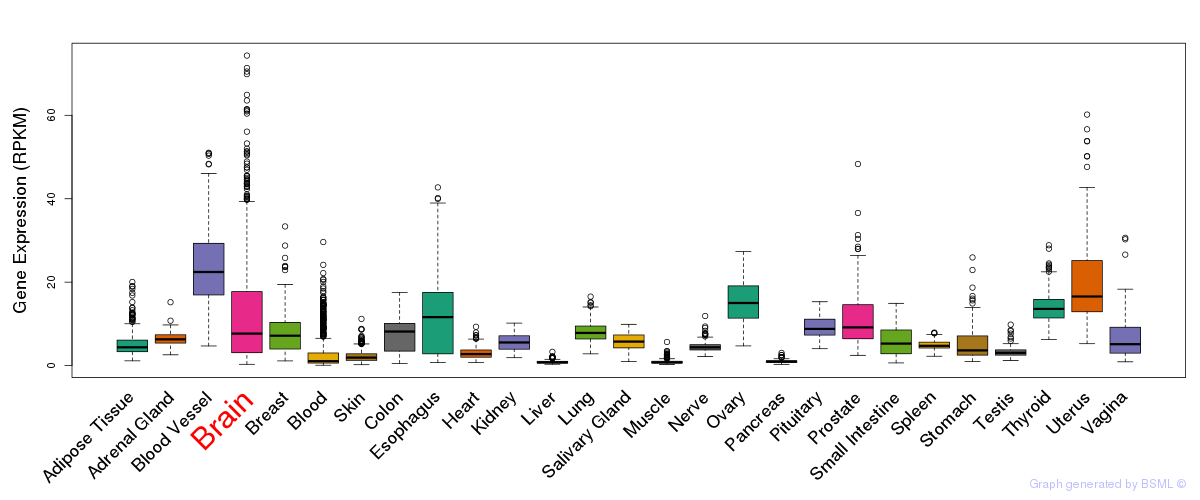
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHCYL1 | DCAL | IRBIT | PRO0233 | XPVKONA | S-adenosylhomocysteine hydrolase-like 1 | - | HPRD,BioGRID | 12525476 |
| BANK1 | BANK | FLJ20706 | FLJ34204 | B-cell scaffold protein with ankyrin repeats 1 | - | HPRD,BioGRID | 11782428 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Affinity Capture-Western | BioGRID | 15613488 |
| CA8 | CA-VIII | CALS | CARP | MGC120502 | MGC99509 | carbonic anhydrase VIII | - | HPRD,BioGRID | 12611586 |
| CABP1 | CALBRAIN | HCALB_BR | calcium binding protein 1 | Affinity Capture-Western in vitro in vivo Reconstituted Complex | BioGRID | 12032348 |14685260 |
| EPB41L1 | 4.1N | DKFZp686H17242 | KIAA0338 | MGC11072 | erythrocyte membrane protein band 4.1-like 1 | - | HPRD,BioGRID | 12444087 |12676536 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | - | HPRD,BioGRID | 9346894 |
| GRM1 | GPRC1A | GRM1A | MGLUR1 | MGLUR1A | mGlu1 | glutamate receptor, metabotropic 1 | - | HPRD,BioGRID | 9808459 |
| HOMER1 | HOMER | HOMER1A | HOMER1B | HOMER1C | SYN47 | Ves-1 | homer homolog 1 (Drosophila) | - | HPRD,BioGRID | 9808459 |
| HOMER2 | ACPD | CPD | HOMER-2 | HOMER2A | HOMER2B | Vesl-2 | homer homolog 2 (Drosophila) | - | HPRD,BioGRID | 9808459 |
| HOMER3 | HOMER-3 | homer homolog 3 (Drosophila) | in vitro in vivo | BioGRID | 9808459 |
| ITPR3 | FLJ36205 | IP3R | IP3R3 | inositol 1,4,5-triphosphate receptor, type 3 | in vivo | BioGRID | 7559486 |
| MRVI1 | IRAG | JAW1L | murine retrovirus integration site 1 homolog | - | HPRD,BioGRID | 10724174 |
| PRKG1 | CGKI | DKFZp686K042 | FLJ36117 | MGC71944 | PGK | PKG | PRKG1B | PRKGR1B | cGKI-BETA | cGKI-alpha | protein kinase, cGMP-dependent, type I | Affinity Capture-Western Biochemical Activity | BioGRID | 10724174 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Affinity Capture-Western Reconstituted Complex | BioGRID | 12766172 |
| SDC2 | HSPG | HSPG1 | SYND2 | syndecan 2 | Affinity Capture-Western | BioGRID | 12676536 |
| STARD13 | DLC2 | FLJ37385 | GT650 | StAR-related lipid transfer (START) domain containing 13 | STARD13 (RhoGAP) interacts with ITPR1. | BIND | 14697242 |
| STARD13 | DLC2 | FLJ37385 | GT650 | StAR-related lipid transfer (START) domain containing 13 | Two-hybrid | BioGRID | 14697242 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC2 | - | transient receptor potential cation channel, subfamily C, member 2 (pseudogene) | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC4 | HTRP4 | MGC119570 | MGC119571 | MGC119572 | MGC119573 | TRP4 | transient receptor potential cation channel, subfamily C, member 4 | Affinity Capture-Western Two-hybrid | BioGRID | 11163362 |14505576 |
| TRPC5 | TRP5 | transient receptor potential cation channel, subfamily C, member 5 | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC6 | FLJ11098 | FLJ14863 | FSGS2 | TRP6 | transient receptor potential cation channel, subfamily C, member 6 | Affinity Capture-Western | BioGRID | 14505576 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| ST WNT CA2 CYCLIC GMP PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST GAQ PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS DN | 68 | 49 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 2 | 42 | 31 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN | 51 | 38 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN D6MIT150 QTL CIS | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN | 50 | 32 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO DISTAL AND PROXIMAL DENDRITES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ WALDENSTROEMS MACROGLOBULINEMIA 1 UP | 9 | 5 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| CHEMELLO SOLEUS VS EDL MYOFIBERS DN | 19 | 10 | All SZGR 2.0 genes in this pathway |