Gene Page: JUND
Summary ?
| GeneID | 3727 |
| Symbol | JUND |
| Synonyms | AP-1 |
| Description | jun D proto-oncogene |
| Reference | MIM:165162|HGNC:HGNC:6206|Ensembl:ENSG00000130522|HPRD:01304|Vega:OTTHUMG00000183355 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.408 |
| Sherlock p-value | 0.15 |
| Fetal beta | -1.16 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
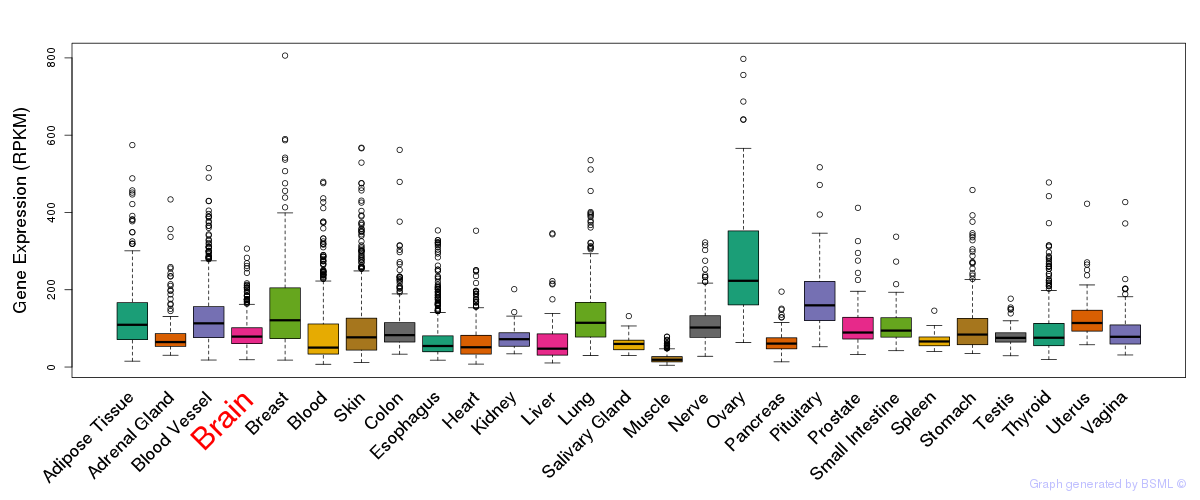
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AAK1 | 0.91 | 0.90 |
| TGOLN2 | 0.90 | 0.92 |
| ARFGEF2 | 0.89 | 0.90 |
| PTPRJ | 0.88 | 0.93 |
| ATP2A2 | 0.88 | 0.89 |
| MFSD6 | 0.88 | 0.88 |
| STS | 0.88 | 0.87 |
| PEG3AS | 0.88 | 0.89 |
| SYT11 | 0.88 | 0.92 |
| SYNJ1 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GNG11 | -0.42 | -0.53 |
| C1orf61 | -0.41 | -0.54 |
| C1orf54 | -0.41 | -0.55 |
| SYCP3 | -0.41 | -0.55 |
| AF347015.21 | -0.41 | -0.48 |
| DBI | -0.40 | -0.50 |
| IL32 | -0.39 | -0.50 |
| C2orf84 | -0.38 | -0.42 |
| RPL31 | -0.38 | -0.36 |
| PFDN5 | -0.37 | -0.31 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATF3 | - | activating transcription factor 3 | - | HPRD,BioGRID | 8152431 |
| BATF | B-ATF | BATF1 | SFA-2 | basic leucine zipper transcription factor, ATF-like | - | HPRD,BioGRID | 8570175 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | - | HPRD,BioGRID | 12165517 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 12080089 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with JunD. | BIND | 12080089 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | - | HPRD,BioGRID | 8837781 |
| CYP19A1 | ARO | ARO1 | CPV1 | CYAR | CYP19 | MGC104309 | P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 | JUND interacts with the CYP19A1 (aromatase PII) promoter. | BIND | 15688015 |
| DDIT3 | CEBPZ | CHOP | CHOP10 | GADD153 | MGC4154 | DNA-damage-inducible transcript 3 | - | HPRD,BioGRID | 10523647 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 12193410 |
| FOSL2 | FLJ23306 | FRA2 | FOS-like antigen 2 | - | HPRD | 10713367 |12052862 |
| JDP2 | JUNDM2 | Jun dimerization protein 2 | - | HPRD | 9154808 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 12052834 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | - | HPRD | 8945519 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | - | HPRD,BioGRID | 9989505 |12226747 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | Menin directly represses JunD-activated transcription | BIND | 10500243 |
| NFE2L1 | FLJ00380 | LCR-F1 | NRF1 | TCF11 | nuclear factor (erythroid-derived 2)-like 1 | - | HPRD | 9872330 |
| NFE2L2 | NRF2 | nuclear factor (erythroid-derived 2)-like 2 | - | HPRD | 9872330 |
| PLEKHO1 | CKIP-1 | OC120 | pleckstrin homology domain containing, family O member 1 | CKIP-1 interacts with JunD. | BIND | 15706351 |
| RFWD2 | COP1 | FLJ10416 | RNF200 | ring finger and WD repeat domain 2 | - | HPRD,BioGRID | 12615916 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Reconstituted Complex | BioGRID | 10220381 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD | 10220381 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD | 7848298 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCELLSURVIVAL PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP | 52 | 38 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE UP | 136 | 80 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK8 AND MAPK9 TARGETS | 8 | 7 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| HUI MAPK14 TARGETS UP | 21 | 11 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 5 UP | 16 | 7 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C6 | 39 | 27 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 4HR | 35 | 28 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G2 | 30 | 18 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS | 14 | 12 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION | 30 | 21 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM RISK UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G24 | 28 | 19 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |