Gene Page: KCNJ4
Summary ?
| GeneID | 3761 |
| Symbol | KCNJ4 |
| Synonyms | HIR|HIRK2|HRK1|IRK-3|IRK3|Kir2.3 |
| Description | potassium voltage-gated channel subfamily J member 4 |
| Reference | MIM:600504|HGNC:HGNC:6265|Ensembl:ENSG00000168135|HPRD:02738|Vega:OTTHUMG00000151131 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.956 |
| Fetal beta | -2.062 |
| Support | EXCITABILITY G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
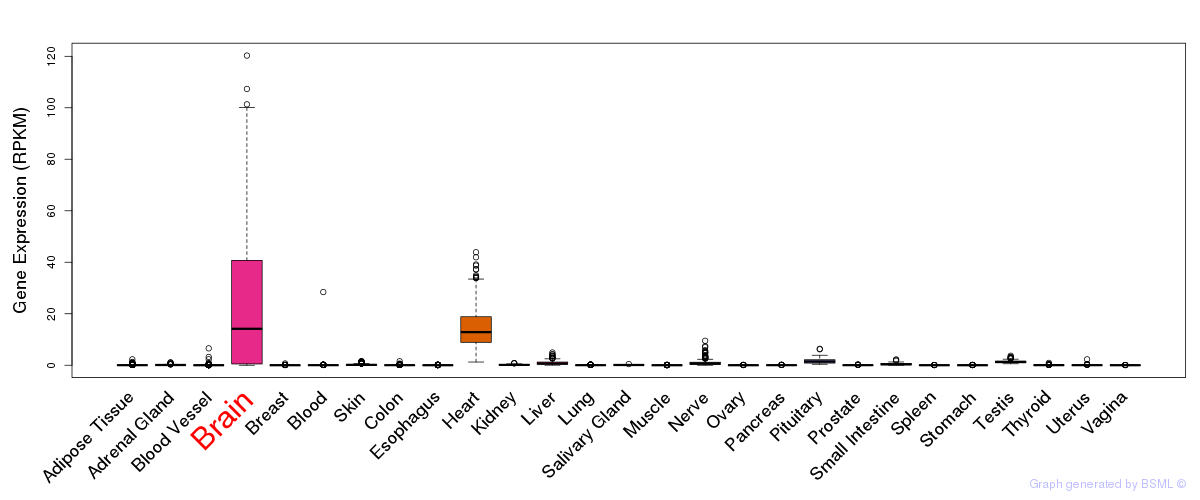
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MYO5A | 0.89 | 0.91 |
| CDKL5 | 0.89 | 0.92 |
| HECW2 | 0.87 | 0.88 |
| PRICKLE2 | 0.87 | 0.88 |
| PCLO | 0.86 | 0.89 |
| INPP4A | 0.86 | 0.88 |
| FAM20B | 0.86 | 0.88 |
| DLG3 | 0.86 | 0.87 |
| MEF2A | 0.86 | 0.87 |
| ASTN1 | 0.85 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.54 | -0.63 |
| AL139819.3 | -0.54 | -0.60 |
| AP002478.3 | -0.53 | -0.61 |
| FXYD1 | -0.53 | -0.55 |
| AF347015.21 | -0.52 | -0.56 |
| ACSF2 | -0.52 | -0.57 |
| HIGD1B | -0.52 | -0.57 |
| AC021016.1 | -0.52 | -0.56 |
| TLCD1 | -0.51 | -0.54 |
| DBI | -0.51 | -0.61 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD | 11742811 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | Affinity Capture-Western | BioGRID | 14960569 |15024025 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11181181 |14960569 |15024025 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | Affinity Capture-Western | BioGRID | 11997254 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 10627592 |11997254 |14960569 |
| DMD | BMD | CMD3B | DXS142 | DXS164 | DXS206 | DXS230 | DXS239 | DXS268 | DXS269 | DXS270 | DXS272 | dystrophin | Affinity Capture-Western | BioGRID | 15024025 |
| GNGT2 | G-GAMMA-8 | G-GAMMA-C | GNG8 | GNG9 | GNGT8 | guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | - | HPRD,BioGRID | 8943291 |
| IL16 | FLJ16806 | FLJ42735 | FLJ44234 | HsT19289 | IL-16 | LCF | prIL-16 | interleukin 16 (lymphocyte chemoattractant factor) | - | HPRD,BioGRID | 10479680 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | Affinity Capture-Western Two-hybrid | BioGRID | 11742811 |
| LIN7C | FLJ11215 | LIN-7-C | LIN-7C | MALS-3 | MALS3 | VELI3 | lin-7 homolog C (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |15024025 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | Affinity Capture-Western | BioGRID | 15024025 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA B RECEPTOR ACTIVATION | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION G1 G2 UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON UP | 77 | 47 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |