Gene Page: KCNJ9
Summary ?
| GeneID | 3765 |
| Symbol | KCNJ9 |
| Synonyms | GIRK3|KIR3.3 |
| Description | potassium voltage-gated channel subfamily J member 9 |
| Reference | MIM:600932|HGNC:HGNC:6270|HPRD:02959| |
| Gene type | protein-coding |
| Map location | 1q23.2 |
| Fetal beta | -2.132 |
| DMG | 1 (# studies) |
| Support | CANABINOID EXCITABILITY SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6703 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16879052 | 1 | 160054270 | KCNJ9 | 4.31E-4 | 0.261 | 0.045 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
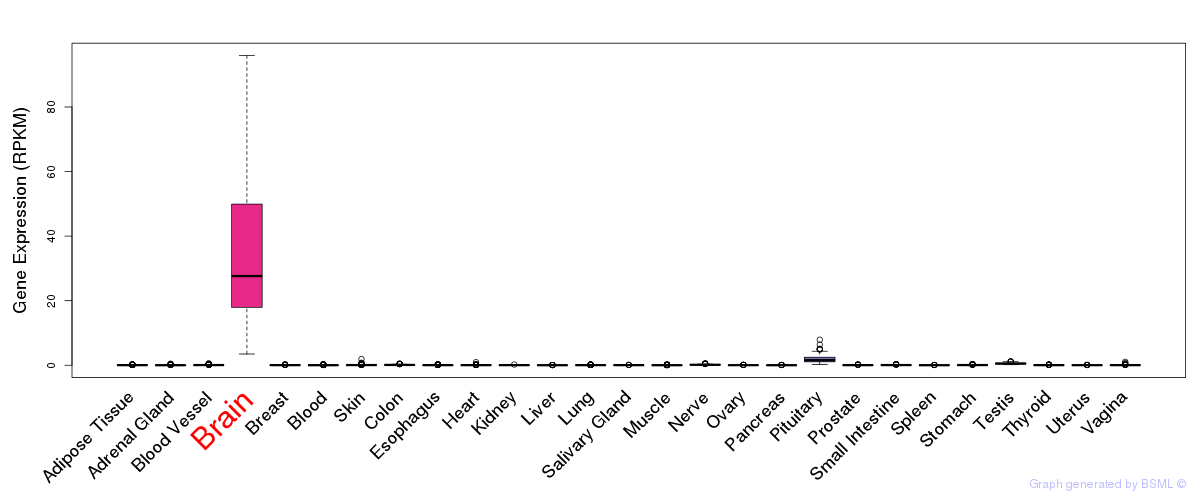
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MICAL2 | 0.87 | 0.79 |
| SNAP25 | 0.86 | 0.78 |
| KCNK1 | 0.86 | 0.86 |
| EXTL1 | 0.85 | 0.87 |
| PIP5K1B | 0.85 | 0.86 |
| CAMK2A | 0.85 | 0.85 |
| VSNL1 | 0.85 | 0.75 |
| KCNAB2 | 0.85 | 0.79 |
| SYT12 | 0.84 | 0.87 |
| VIPR1 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.59 | -0.55 |
| SH3BP2 | -0.58 | -0.67 |
| TUBB2B | -0.58 | -0.63 |
| BCL7C | -0.57 | -0.70 |
| SH2B2 | -0.56 | -0.65 |
| PKN1 | -0.56 | -0.56 |
| TRAF4 | -0.56 | -0.65 |
| PDE9A | -0.55 | -0.61 |
| C21orf30 | -0.54 | -0.54 |
| FNBP1L | -0.54 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0015467 | G-protein activated inward rectifier potassium channel activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA B RECEPTOR ACTIVATION | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL CIS | 65 | 38 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST GENETIC VARIANCE | 37 | 21 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K27ME3 | 54 | 32 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K27ME3 | 42 | 27 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |