Gene Page: RBM12B
Summary ?
| GeneID | 389677 |
| Symbol | RBM12B |
| Synonyms | MGC:33837 |
| Description | RNA binding motif protein 12B |
| Reference | HGNC:HGNC:32310|HPRD:14213| |
| Gene type | protein-coding |
| Map location | 8q22.1 |
| Pascal p-value | 0.308 |
| Fetal beta | 0.809 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27481254 | 8 | 94753071 | RBM12B | 1.55E-9 | -0.019 | 1.44E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | RBM12B | 389677 | 6.996E-4 | trans | ||
| rs7584986 | chr2 | 184111432 | RBM12B | 389677 | 7.845E-4 | trans | ||
| rs10183246 | chr2 | 237187882 | RBM12B | 389677 | 0.19 | trans | ||
| rs1155107 | chr3 | 26843376 | RBM12B | 389677 | 0.02 | trans | ||
| rs11186599 | chr10 | 93285701 | RBM12B | 389677 | 0.19 | trans | ||
| rs7076439 | chr10 | 93286841 | RBM12B | 389677 | 0.01 | trans | ||
| rs11186601 | chr10 | 93287199 | RBM12B | 389677 | 0.01 | trans | ||
| rs4980232 | chr10 | 124577756 | RBM12B | 389677 | 0.08 | trans | ||
| rs1324669 | chr13 | 107872446 | RBM12B | 389677 | 8.902E-4 | trans | ||
| rs16955618 | chr15 | 29937543 | RBM12B | 389677 | 0.02 | trans | ||
| rs6098023 | chr20 | 53113740 | RBM12B | 389677 | 0.08 | trans | ||
| rs17145698 | chrX | 40218345 | RBM12B | 389677 | 0.15 | trans |
Section II. Transcriptome annotation
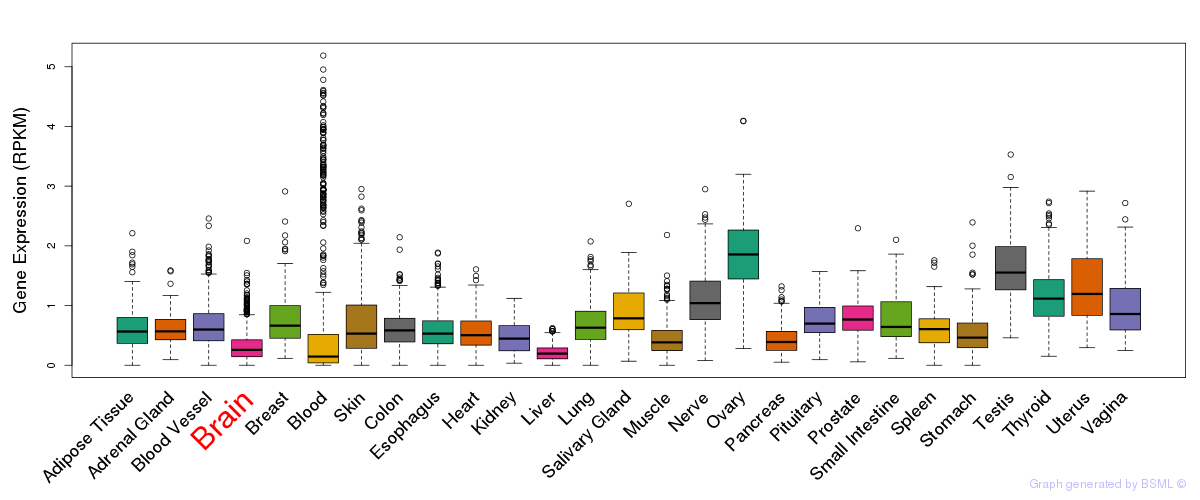
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS INDUCED BY AKT1 48HR DN | 27 | 17 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS RESPONSIVE TO ESTROGEN UP | 31 | 20 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |