Gene Page: RHOG
Summary ?
| GeneID | 391 |
| Symbol | RHOG |
| Synonyms | ARHG |
| Description | ras homolog family member G |
| Reference | MIM:179505|HGNC:HGNC:672|Ensembl:ENSG00000177105|HPRD:01537|Vega:OTTHUMG00000012287 |
| Gene type | protein-coding |
| Map location | 11p15.5-p15.4 |
| Pascal p-value | 0.332 |
| Sherlock p-value | 0.041 |
| Fetal beta | -1.086 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12991791 | 11 | 3863056 | RHOG | 1.37E-9 | -0.02 | 1.37E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7830259 | chr8 | 14087643 | RHOG | 391 | 0.04 | trans | ||
| rs6632306 | chrX | 35590460 | RHOG | 391 | 0.06 | trans |
Section II. Transcriptome annotation
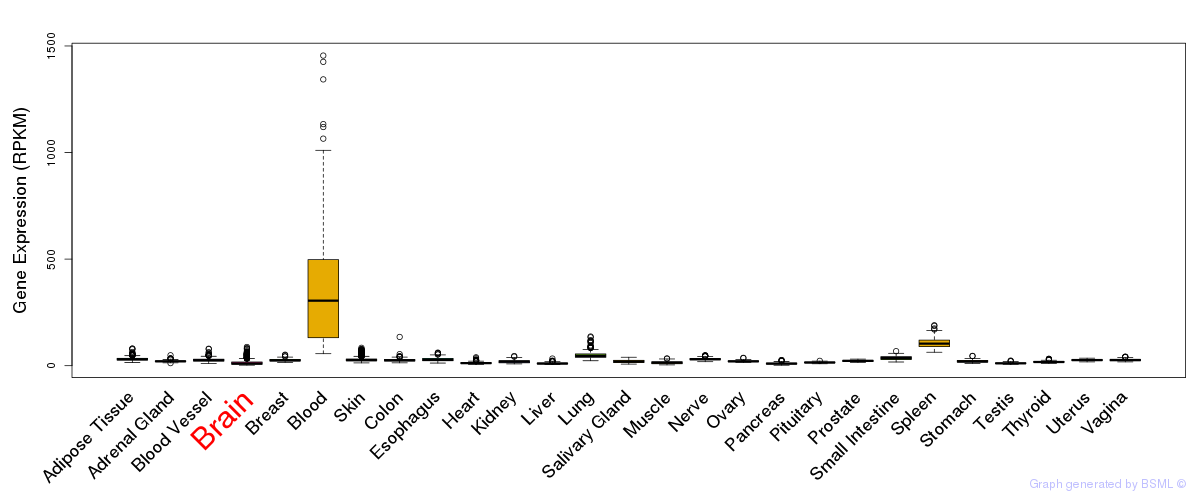
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACAP2 | 0.87 | 0.88 |
| LIFR | 0.86 | 0.87 |
| PTAR1 | 0.86 | 0.85 |
| DNAJC3 | 0.85 | 0.86 |
| PIK3C2A | 0.85 | 0.87 |
| STK38L | 0.85 | 0.84 |
| ITGAV | 0.84 | 0.83 |
| DPY19L4 | 0.84 | 0.85 |
| KIAA1033 | 0.84 | 0.82 |
| MKLN1 | 0.84 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL022328.1 | -0.46 | -0.47 |
| IL32 | -0.45 | -0.43 |
| HES4 | -0.43 | -0.46 |
| MRPL52 | -0.42 | -0.38 |
| NUDT8 | -0.41 | -0.45 |
| AC016757.1 | -0.41 | -0.39 |
| AF347015.21 | -0.41 | -0.30 |
| DPM3 | -0.40 | -0.43 |
| APOC1 | -0.40 | -0.33 |
| ST20 | -0.40 | -0.37 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGDIG | RHOGDI-3 | Rho GDP dissociation inhibitor (GDI) gamma | - | HPRD,BioGRID | 8939998 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | - | HPRD,BioGRID | 12879077 |
| ECT2 | FLJ10461 | MGC138291 | epithelial cell transforming sequence 2 oncogene | - | HPRD | 12376551 |
| ELMO1 | CED-12 | CED12 | ELMO-1 | KIAA0281 | MGC126406 | engulfment and cell motility 1 | ELMO1 interacts with RhoG. | BIND | 15620647 |
| ELMO2 | CED-12 | CED12 | ELMO-2 | FLJ11656 | KIAA1834 | engulfment and cell motility 2 | - | HPRD | 12879077 |
| ELMOD2 | 9830169G11Rik | MGC10084 | ELMO/CED-12 domain containing 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12879077 |
| IQGAP2 | - | IQ motif containing GTPase activating protein 2 | Reconstituted Complex | BioGRID | 12376551 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | - | HPRD,BioGRID | 11689693 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | - | HPRD,BioGRID | 12376551 |
| MCF2L | ARHGEF14 | DBS | FLJ12122 | KIAA0362 | OST | MCF.2 cell line derived transforming sequence-like | - | HPRD | 12376551 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD | 12376551 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD | 11909943 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10523675 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| HUMMERICH BENIGN SKIN TUMOR UP | 18 | 8 | All SZGR 2.0 genes in this pathway |
| HUMMERICH MALIGNANT SKIN TUMOR UP | 16 | 8 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ITO PTTG1 TARGETS DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |