| KEGG FOCAL ADHESION
| 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION
| 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER
| 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER
| 84 | 67 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY
| 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN4 PATHWAY
| 11 | 5 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY
| 46 | 29 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY
| 46 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS
| 79 | 48 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP
| 783 | 507 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP
| 276 | 187 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP
| 430 | 232 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN
| 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN
| 281 | 186 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP
| 112 | 68 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN
| 240 | 171 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN
| 260 | 143 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A DN
| 90 | 61 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP
| 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY DN
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS
| 539 | 324 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP
| 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP
| 487 | 286 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON
| 149 | 76 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP
| 56 | 34 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP
| 60 | 40 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS
| 318 | 215 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN
| 129 | 86 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN
| 162 | 102 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN
| 86 | 67 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS
| 191 | 131 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 DN
| 52 | 35 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM
| 302 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP
| 180 | 125 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP
| 178 | 111 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN
| 287 | 208 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN
| 142 | 94 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN
| 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN
| 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN
| 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN
| 270 | 181 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL
| 110 | 68 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN
| 145 | 93 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP
| 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN
| 138 | 92 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN
| 366 | 257 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP
| 298 | 174 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN
| 442 | 275 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP
| 105 | 75 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION
| 368 | 247 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN
| 207 | 139 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D
| 280 | 158 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS
| 32 | 22 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP
| 169 | 105 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP
| 116 | 65 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN
| 214 | 124 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS
| 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME
| 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA BASEMENT MEMBRANES
| 40 | 22 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME
| 1028 | 559 | All SZGR 2.0 genes in this pathway |
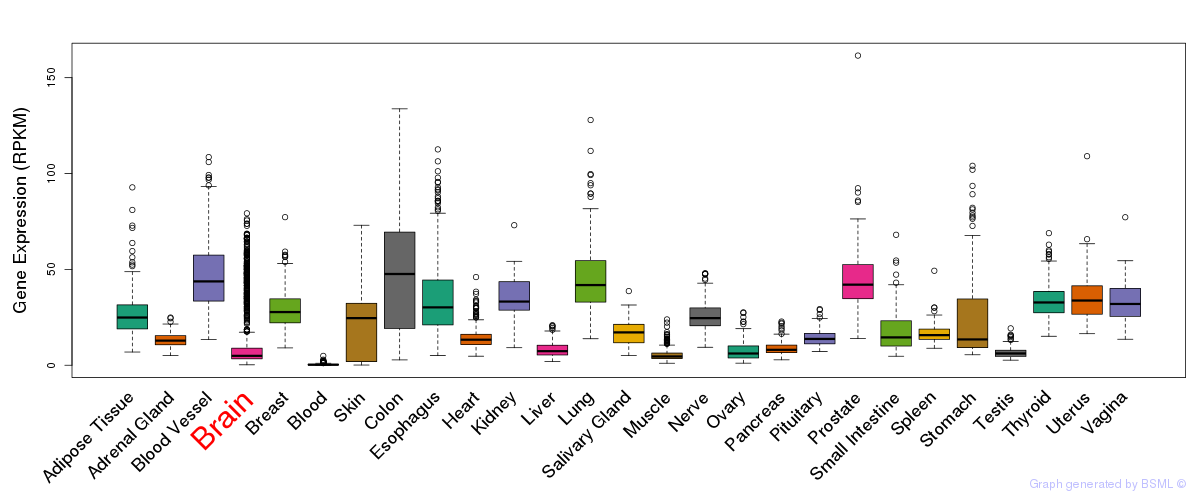
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation