Gene Page: LBP
Summary ?
| GeneID | 3929 |
| Symbol | LBP |
| Synonyms | BPIFD2 |
| Description | lipopolysaccharide binding protein |
| Reference | MIM:151990|HGNC:HGNC:6517|Ensembl:ENSG00000129988|HPRD:07175|Vega:OTTHUMG00000032447 |
| Gene type | protein-coding |
| Map location | 20q11.23 |
| Pascal p-value | 0.826 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
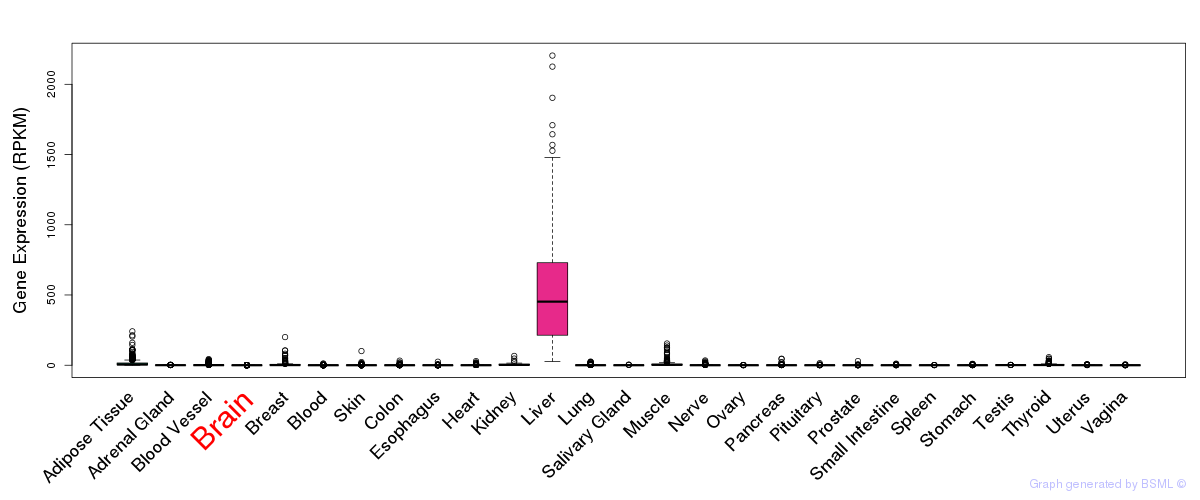
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TGFBRAP1 | 0.95 | 0.95 |
| AC097382.4 | 0.94 | 0.94 |
| DHX37 | 0.94 | 0.95 |
| SREBF2 | 0.94 | 0.95 |
| NEURL4 | 0.93 | 0.95 |
| CLIP2 | 0.93 | 0.94 |
| DIP2C | 0.93 | 0.94 |
| FOXK2 | 0.93 | 0.95 |
| KIAA1967 | 0.93 | 0.94 |
| BICD2 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.86 |
| MT-CO2 | -0.77 | -0.86 |
| AF347015.27 | -0.76 | -0.84 |
| AF347015.21 | -0.75 | -0.91 |
| AF347015.33 | -0.74 | -0.81 |
| AC021016.1 | -0.74 | -0.82 |
| FXYD1 | -0.73 | -0.80 |
| AF347015.8 | -0.73 | -0.84 |
| MT-CYB | -0.73 | -0.81 |
| HIGD1B | -0.73 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | ISS | Neurotransmitter (GO term level: 4) | 2477488 |
| GO:0001530 | lipopolysaccharide binding | IEA | - | |
| GO:0008289 | lipid binding | IEA | - | |
| GO:0043498 | cell surface binding | ISS | 2477488 | |
| GO:0051636 | Gram-negative bacterial cell surface binding | IDA | 11528597 | |
| GO:0051636 | Gram-negative bacterial cell surface binding | ISS | 2402637 | |
| GO:0051637 | Gram-positive bacterial cell surface binding | IDA | 12932360 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002281 | macrophage activation during immune response | ISS | 2402637 | |
| GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway | IDA | 11528597 | |
| GO:0006968 | cellular defense response | ISS | 2402637 | |
| GO:0006953 | acute-phase response | IEP | 12932360 | |
| GO:0006953 | acute-phase response | ISS | 2477488 | |
| GO:0006869 | lipid transport | IEA | - | |
| GO:0008228 | opsonization | IC | 12932360 | |
| GO:0008228 | opsonization | ISS | 2477488 | |
| GO:0015920 | lipopolysaccharide transport | IDA | 8985160 | |
| GO:0032757 | positive regulation of interleukin-8 production | IDA | 8409400 | |
| GO:0032496 | response to lipopolysaccharide | IDA | 8409400 |11739189 | |
| GO:0032490 | detection of molecule of bacterial origin | IDA | 11528597 | |
| GO:0032760 | positive regulation of tumor necrosis factor production | IDA | 8409400 | |
| GO:0032760 | positive regulation of tumor necrosis factor production | ISS | 2402637 | |
| GO:0032755 | positive regulation of interleukin-6 production | IDA | 8409400 | |
| GO:0032720 | negative regulation of tumor necrosis factor production | IDA | 11739189 | |
| GO:0031663 | lipopolysaccharide-mediated signaling pathway | IDA | 8409400 |11739189 | |
| GO:0043032 | positive regulation of macrophage activation | IDA | 8409400 | |
| GO:0050830 | defense response to Gram-positive bacterium | IC | 12932360 | |
| GO:0050830 | defense response to Gram-positive bacterium | IEP | 12932360 | |
| GO:0050829 | defense response to Gram-negative bacterium | ISS | 2402637 | |
| GO:0045087 | innate immune response | IC | 12932360 | |
| GO:0045087 | innate immune response | ISS | 2477488 | |
| GO:0060265 | positive regulation of respiratory burst during acute inflammatory response | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2477488 |9665271 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 2402637 |11528597 | |
| GO:0005615 | extracellular space | ISS | 2477488 | |
| GO:0005634 | nucleus | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 UP | 46 | 29 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 DN | 78 | 44 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID CEBPA NETWORK | 28 | 19 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 DN | 50 | 24 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL PROGENITOR UP | 58 | 30 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-500 | 162 | 168 | m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.