Gene Page: ARHGDIA
Summary ?
| GeneID | 396 |
| Symbol | ARHGDIA |
| Synonyms | GDIA1|HEL-S-47e|NPHS8|RHOGDI|RHOGDI-1 |
| Description | Rho GDP dissociation inhibitor (GDI) alpha |
| Reference | MIM:601925|HGNC:HGNC:678|Ensembl:ENSG00000141522|HPRD:03565|Vega:OTTHUMG00000178352 |
| Gene type | protein-coding |
| Map location | 17q25.3 |
| Pascal p-value | 0.799 |
| Sherlock p-value | 0.342 |
| Fetal beta | -0.63 |
| Support | EXOCYTOSIS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
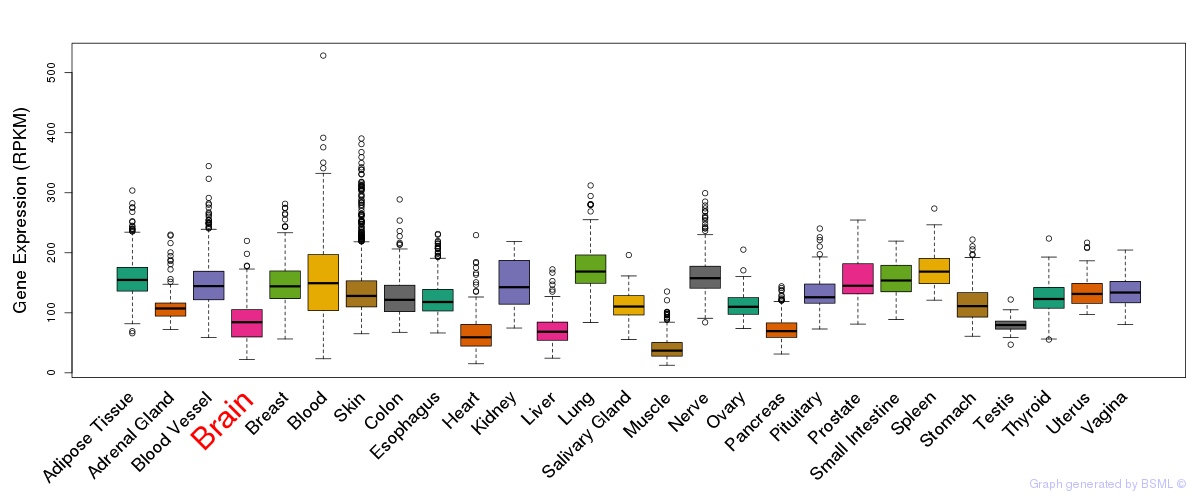
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1QA | 0.49 | 0.41 |
| C1QB | 0.49 | 0.44 |
| TYROBP | 0.48 | 0.41 |
| SERPINA1 | 0.47 | 0.40 |
| C1QC | 0.47 | 0.41 |
| FCER1G | 0.46 | 0.40 |
| GPR34 | 0.46 | 0.36 |
| RGS10 | 0.46 | 0.43 |
| NAPSB | 0.44 | 0.33 |
| PLN | 0.44 | 0.23 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SRGAP1 | -0.27 | -0.24 |
| EIF2C4 | -0.26 | -0.22 |
| SCUBE1 | -0.26 | -0.28 |
| ATXN7L1 | -0.26 | -0.24 |
| MYCN | -0.26 | -0.24 |
| GJC1 | -0.26 | -0.23 |
| ZEB2 | -0.26 | -0.24 |
| AL033532.1 | -0.26 | -0.25 |
| BCORL1 | -0.26 | -0.26 |
| KIAA1949 | -0.26 | -0.23 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | Two-hybrid | BioGRID | 16169070 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Reconstituted Complex | BioGRID | 11513578 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD | 10676816 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | Affinity Capture-MS | BioGRID | 17353931 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Two-hybrid | BioGRID | 16169070 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF2B1 | EIF-2B | EIF-2Balpha | EIF2B | EIF2BA | MGC117409 | MGC125868 | MGC125869 | eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa | Affinity Capture-MS | BioGRID | 17353931 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Co-purification | BioGRID | 15659383 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | Two-hybrid | BioGRID | 16169070 |
| HNRNPA3 | 2610510D13Rik | D10S102 | FBRNP | HNRPA3 | MGC138232 | MGC142030 | heterogeneous nuclear ribonucleoprotein A3 | Affinity Capture-MS | BioGRID | 17353931 |
| HNRNPAB | ABBP1 | FLJ40338 | HNRPAB | heterogeneous nuclear ribonucleoprotein A/B | Affinity Capture-MS | BioGRID | 17353931 |
| IGF2BP1 | CRD-BP | CRDBP | IMP-1 | IMP1 | VICKZ1 | ZBP1 | insulin-like growth factor 2 mRNA binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Two-hybrid | BioGRID | 16169070 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| NEDD4L | FLJ33870 | KIAA0439 | NEDD4-2 | RSP5 | hNedd4-2 | neural precursor cell expressed, developmentally down-regulated 4-like | Two-hybrid | BioGRID | 16169070 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Pak1 interacts with and phosphorylates RhoGDI. This interaction was modeled on a demonstrated interaction between Pak1 from an unspecified species and human RhoGDI. | BIND | 15225553 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD | 10346909 |10673424|11513579 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | - | HPRD,BioGRID | 10673424 |
| RAC2 | EN-7 | Gx | HSPC022 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | - | HPRD | 11513578 |
| RDX | DFNB24 | radixin | - | HPRD,BioGRID | 9287351 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | - | HPRD,BioGRID | 11149925 |
| RHOC | ARH9 | ARHC | H9 | MGC1448 | MGC61427 | RHOH9 | ras homolog gene family, member C | Affinity Capture-MS | BioGRID | 17353931 |
| RHOH | ARHH | TTF | ras homolog gene family, member H | Affinity Capture-Western | BioGRID | 11809807 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | Two-hybrid | BioGRID | 16169070 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Co-purification | BioGRID | 15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G4 | 21 | 15 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| ITO PTTG1 TARGETS DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| LOPEZ TRANSLATION VIA FN1 SIGNALING | 35 | 21 | All SZGR 2.0 genes in this pathway |