Gene Page: LIG3
Summary ?
| GeneID | 3980 |
| Symbol | LIG3 |
| Synonyms | LIG2 |
| Description | ligase III, DNA, ATP-dependent |
| Reference | MIM:600940|HGNC:HGNC:6600|Ensembl:ENSG00000005156|HPRD:02966|Vega:OTTHUMG00000128519 |
| Gene type | protein-coding |
| Map location | 17q11.2-q12 |
| Pascal p-value | 0.104 |
| Sherlock p-value | 0.009 |
| Fetal beta | 0.423 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12945428 | 17 | 33307586 | LIG3 | ENSG00000005156.7 | 1.82E-6 | 0 | 73 | gtex_brain_putamen_basal |
| rs3135967 | 17 | 33313729 | LIG3 | ENSG00000005156.7 | 2.62E-7 | 0 | 6216 | gtex_brain_putamen_basal |
| rs2074518 | 17 | 33324382 | LIG3 | ENSG00000005156.7 | 3.165E-7 | 0 | 16869 | gtex_brain_putamen_basal |
| rs1052536 | 17 | 33331575 | LIG3 | ENSG00000005156.7 | 3.165E-7 | 0 | 24062 | gtex_brain_putamen_basal |
| rs1003918 | 17 | 33332177 | LIG3 | ENSG00000005156.7 | 5.073E-7 | 0 | 24664 | gtex_brain_putamen_basal |
| rs12948362 | 17 | 33332629 | LIG3 | ENSG00000005156.7 | 3.165E-7 | 0 | 25116 | gtex_brain_putamen_basal |
| rs10853174 | 17 | 33351917 | LIG3 | ENSG00000005156.7 | 3.165E-7 | 0 | 44404 | gtex_brain_putamen_basal |
| rs1634802 | 17 | 33360374 | LIG3 | ENSG00000005156.7 | 2.007E-7 | 0 | 52861 | gtex_brain_putamen_basal |
| rs810042 | 17 | 33382205 | LIG3 | ENSG00000005156.7 | 3.163E-7 | 0 | 74692 | gtex_brain_putamen_basal |
| rs2339123 | 17 | 33382734 | LIG3 | ENSG00000005156.7 | 3.163E-7 | 0 | 75221 | gtex_brain_putamen_basal |
| rs2339122 | 17 | 33382801 | LIG3 | ENSG00000005156.7 | 3.163E-7 | 0 | 75288 | gtex_brain_putamen_basal |
| rs1634800 | 17 | 33383030 | LIG3 | ENSG00000005156.7 | 3.163E-7 | 0 | 75517 | gtex_brain_putamen_basal |
| rs1088450 | 17 | 33399745 | LIG3 | ENSG00000005156.7 | 6.986E-8 | 0 | 92232 | gtex_brain_putamen_basal |
| rs797990 | 17 | 33412739 | LIG3 | ENSG00000005156.7 | 1.463E-7 | 0 | 105226 | gtex_brain_putamen_basal |
| rs797989 | 17 | 33414758 | LIG3 | ENSG00000005156.7 | 1.084E-7 | 0 | 107245 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
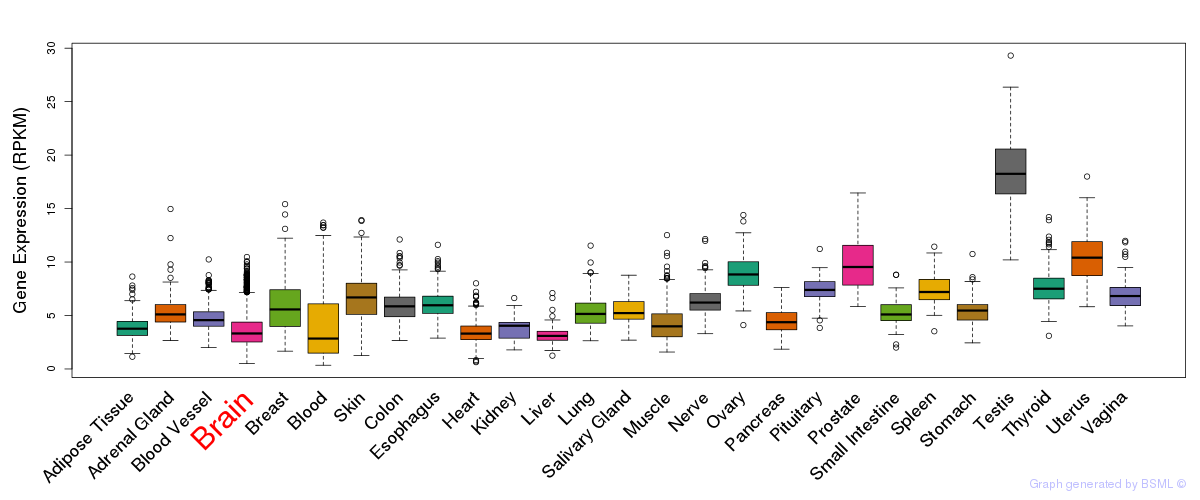
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ITGB5 | 0.72 | 0.69 |
| APCDD1 | 0.70 | 0.66 |
| SLC12A4 | 0.70 | 0.61 |
| LMCD1 | 0.70 | 0.76 |
| GIMAP1 | 0.70 | 0.67 |
| LSR | 0.69 | 0.68 |
| FZD6 | 0.69 | 0.61 |
| A2ML1 | 0.67 | 0.60 |
| BCAN | 0.66 | 0.62 |
| TMEM51 | 0.66 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPR22 | -0.42 | -0.41 |
| C6orf115 | -0.40 | -0.39 |
| GPR21 | -0.40 | -0.41 |
| RLBP1L1 | -0.40 | -0.36 |
| C18orf8 | -0.40 | -0.38 |
| NELL2 | -0.39 | -0.38 |
| NEUROD6 | -0.39 | -0.38 |
| TIAM2 | -0.39 | -0.39 |
| WASF1 | -0.39 | -0.34 |
| NFIL3 | -0.39 | -0.37 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASE EXCISION REPAIR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME BASE EXCISION REPAIR | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |