Gene Page: LPO
Summary ?
| GeneID | 4025 |
| Symbol | LPO |
| Synonyms | SPO |
| Description | lactoperoxidase |
| Reference | MIM:150205|HGNC:HGNC:6678|Ensembl:ENSG00000167419|HPRD:11825|Vega:OTTHUMG00000178921 |
| Gene type | protein-coding |
| Map location | 17q23.1 |
| Pascal p-value | 0.008 |
| Fetal beta | -0.368 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24489015 | 17 | 56316162 | LPO | 9.34E-6 | -0.006 | 0.043 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2190759 | chr17 | 55794698 | LPO | 4025 | 0.13 | cis |
Section II. Transcriptome annotation
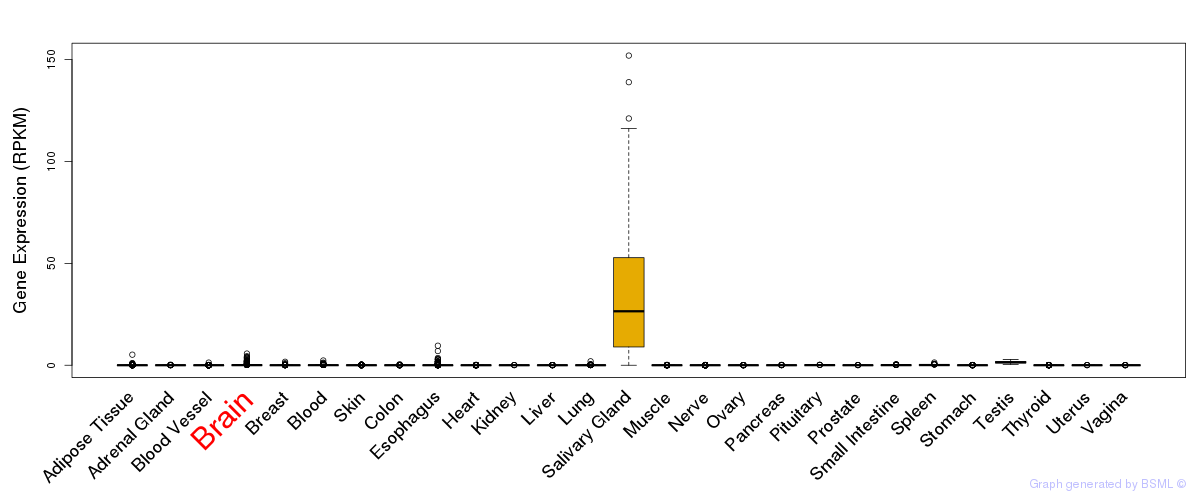
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DPP7 | 0.57 | 0.59 |
| SLC16A11 | 0.57 | 0.59 |
| SELM | 0.56 | 0.53 |
| CHCHD10 | 0.55 | 0.54 |
| KB-1839H6.1 | 0.55 | 0.54 |
| POLD4 | 0.55 | 0.54 |
| C16orf14 | 0.54 | 0.54 |
| NENF | 0.54 | 0.54 |
| DEGS2 | 0.54 | 0.61 |
| CISD3 | 0.53 | 0.54 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.46 | -0.52 |
| RSBN1L | -0.46 | -0.52 |
| ZNF714 | -0.45 | -0.49 |
| ZNF286 | -0.45 | -0.49 |
| ARID4A | -0.45 | -0.51 |
| WHSC1L1 | -0.45 | -0.49 |
| ZNF197 | -0.44 | -0.46 |
| ZNF454 | -0.44 | -0.48 |
| ANKRD12 | -0.44 | -0.53 |
| ZNF616 | -0.44 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND RARA PLZF FUSION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K27ME3 | 74 | 46 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |