| PID P38 MK2 PATHWAY
| 21 | 19 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR MARKERS DN
| 20 | 14 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP
| 473 | 314 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP
| 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP
| 175 | 108 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN
| 260 | 143 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN
| 133 | 77 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP
| 120 | 73 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP
| 68 | 41 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN
| 203 | 134 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP
| 108 | 66 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND
| 69 | 40 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP
| 390 | 236 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN
| 514 | 330 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN
| 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS
| 775 | 494 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN
| 77 | 49 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN
| 309 | 206 | All SZGR 2.0 genes in this pathway |
| FERRANDO LYL1 NEIGHBORS
| 15 | 12 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN
| 366 | 238 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS
| 44 | 31 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN
| 77 | 48 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN
| 225 | 163 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN
| 245 | 144 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP
| 428 | 266 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP
| 253 | 147 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP
| 282 | 183 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP
| 299 | 167 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN
| 546 | 362 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP
| 259 | 185 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN
| 102 | 65 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN
| 50 | 35 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION
| 30 | 21 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN
| 72 | 47 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3
| 128 | 68 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS DN
| 46 | 26 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY DN
| 14 | 11 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1
| 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP
| 84 | 61 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP
| 194 | 133 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF
| 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS DN
| 24 | 16 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE
| 293 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
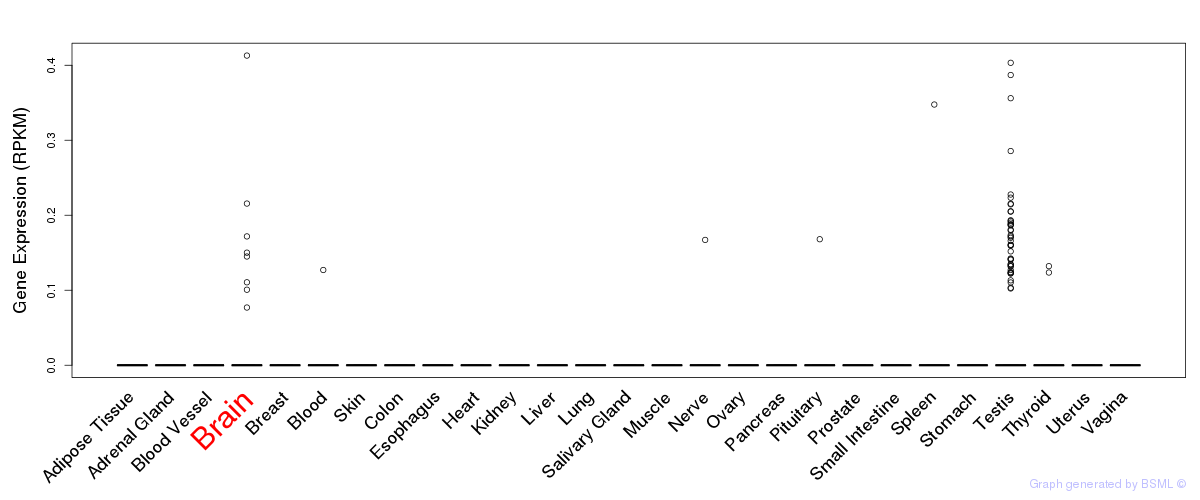
 Differentially methylated gene
Differentially methylated gene