Gene Page: LY6H
Summary ?
| GeneID | 4062 |
| Symbol | LY6H |
| Synonyms | NMLY6 |
| Description | lymphocyte antigen 6 complex, locus H |
| Reference | MIM:603625|HGNC:HGNC:6728|Ensembl:ENSG00000176956|HPRD:04691|Vega:OTTHUMG00000154890 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.937 |
| Fetal beta | -0.376 |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
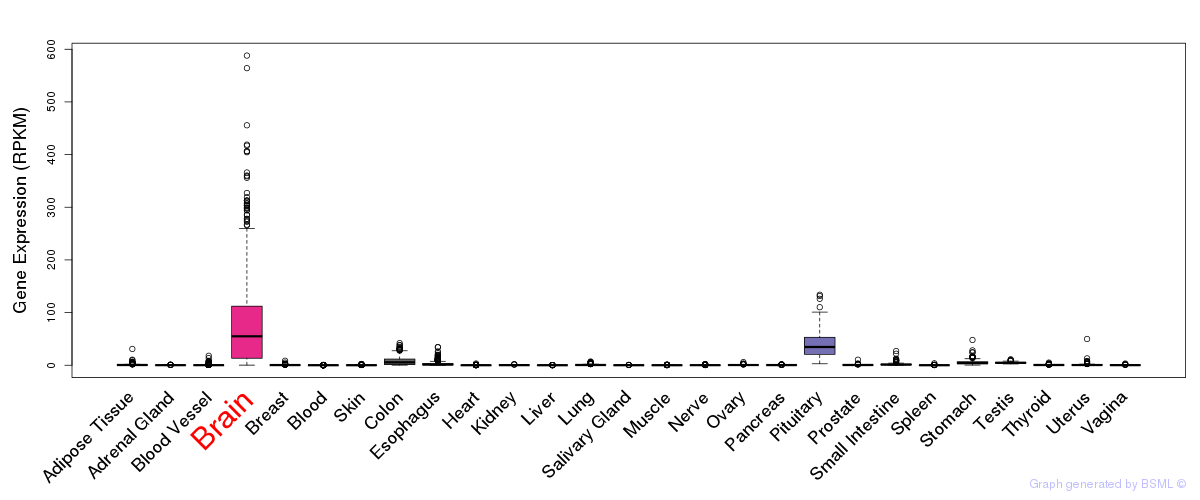
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RUFY3 | 0.92 | 0.94 |
| VEZT | 0.92 | 0.93 |
| OGFOD1 | 0.92 | 0.92 |
| STRN3 | 0.92 | 0.92 |
| GSPT1 | 0.91 | 0.94 |
| THUMPD3 | 0.91 | 0.92 |
| DHX36 | 0.91 | 0.92 |
| ZFP90 | 0.91 | 0.93 |
| DIS3 | 0.91 | 0.92 |
| KPNA3 | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.81 | -0.84 |
| AF347015.8 | -0.78 | -0.84 |
| AF347015.31 | -0.78 | -0.83 |
| AF347015.27 | -0.77 | -0.81 |
| AF347015.33 | -0.77 | -0.81 |
| FXYD1 | -0.77 | -0.83 |
| MT-CYB | -0.76 | -0.81 |
| AF347015.21 | -0.76 | -0.82 |
| IFI27 | -0.76 | -0.82 |
| AF347015.15 | -0.74 | -0.81 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9799603 |
| GO:0009887 | organ morphogenesis | TAS | 9799603 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |