Gene Page: MAD2L1
Summary ?
| GeneID | 4085 |
| Symbol | MAD2L1 |
| Synonyms | HSMAD2|MAD2 |
| Description | MAD2 mitotic arrest deficient-like 1 (yeast) |
| Reference | MIM:601467|HGNC:HGNC:6763|HPRD:03274| |
| Gene type | protein-coding |
| Map location | 4q27 |
| Pascal p-value | 0.155 |
| Fetal beta | 0.735 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23781003 | 4 | 120988207 | MAD2L1 | 6.64E-9 | -0.024 | 3.41E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
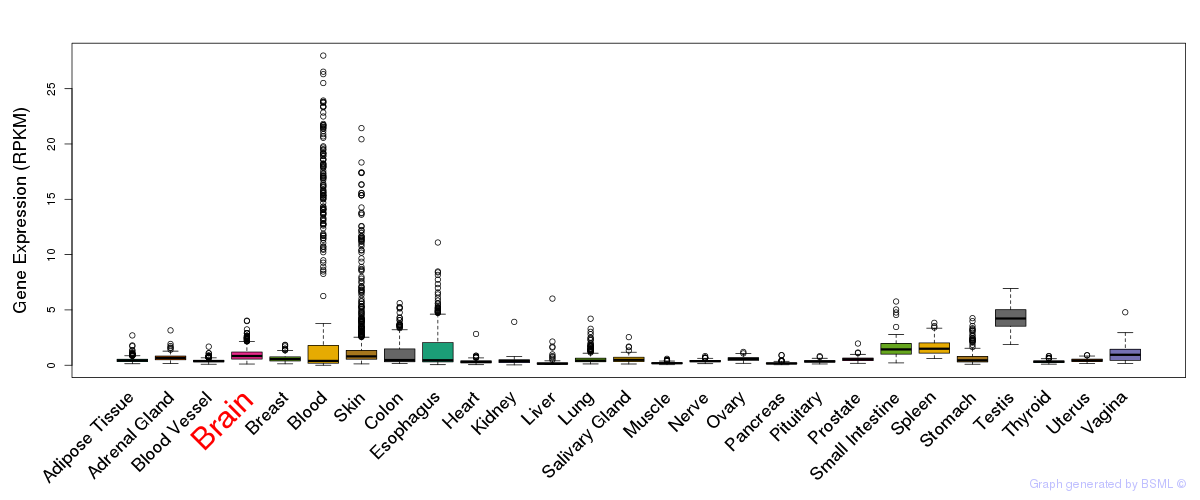
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAM10 | AD10 | CD156c | HsT18717 | MADM | kuz | ADAM metallopeptidase domain 10 | Affinity Capture-Western | BioGRID | 11741929 |
| ADAM15 | MDC15 | ADAM metallopeptidase domain 15 | Affinity Capture-Western | BioGRID | 11741929 |
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | MAD2 interacts with TACE. | BIND | 10527948 |
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | - | HPRD,BioGRID | 10527948 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | - | HPRD,BioGRID | 9637688 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | Affinity Capture-Western Co-purification | BioGRID | 11535616 |
| CDC16 | APC6 | cell division cycle 16 homolog (S. cerevisiae) | - | HPRD | 9628895 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | - | HPRD | 9628895 |9637688 |9736712 |10700282 |11804586 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | MAD2 interacts with p55CDC. | BIND | 12456649 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | MAD2L1 (Mad2) interacts with Cdc20. | BIND | 15694304 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | Mad2 interacts with Cdc20. | BIND | 15525512 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | Affinity Capture-Western Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 9628895 |9637688 |9736712 |10700282 |11438673 |11707408 |12196507 |14561775 |14607335 |
| CDC27 | APC3 | CDC27Hs | D0S1430E | D17S978E | HNUC | cell division cycle 27 homolog (S. cerevisiae) | - | HPRD,BioGRID | 9736712 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD,BioGRID | 11551900 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | MAD2 interacts with CSF2RB. This interaction was modelled on a demonstrated interaction between mouse MAD2 and human CSFR2B. | BIND | 11551900 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F1 interacts with the MAD2 promoter. | BIND | 11799067 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F interacts with the MAD2 promoter and 5-prime UTR. | BIND | 15306814 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with the MAD2 promoter region. | BIND | 11799067 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | MAD2 interacts with ER beta. This interaction was modeled on a demonstrated interaction between MAD2 from human and sheep and ER beta from rat and mouse. | BIND | 10706629 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD,BioGRID | 10706629 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | - | HPRD | 11804586 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | MAD1L1 (Mad1) interacts with MAD2L1 (Mad2). | BIND | 15694304 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | Reconstituted Complex Two-hybrid | BioGRID | 10660610 |11707408 |16189514 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | TXBP181 interacts with HsMAD2. | BIND | 9546394 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | MAD2 interacts with CMT2. | BIND | 12456649 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | - | HPRD,BioGRID | 12456649 |
| MAD2L2 | MAD2B | REV7 | MAD2 mitotic arrest deficient-like 2 (yeast) | Reconstituted Complex | BioGRID | 10660610 |
| MBL2 | COLEC1 | HSMBPC | MBL | MBP | MBP1 | MGC116832 | MGC116833 | mannose-binding lectin (protein C) 2, soluble (opsonic defect) | - | HPRD | 11804586 |
| NDC80 | HEC | HEC1 | KNTC2 | TID3 | hsNDC80 | NDC80 homolog, kinetochore complex component (S. cerevisiae) | - | HPRD | 12351790 |
| REV3L | POLZ | REV3 | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | - | HPRD,BioGRID | 10660610 |
| REV3L | POLZ | REV3 | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | hREV3 interacts with hMAD2. | BIND | 10660610 |
| RIPK5 | DustyPK | HDCMD38P | KIAA0472 | RIP5 | receptor interacting protein kinase 5 | Affinity Capture-MS | BioGRID | 17353931 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| UBD | FAT10 | GABBR1 | UBD-3 | ubiquitin D | MAD2 interacts with FAT10. | BIND | 10200259 |
| UBD | FAT10 | GABBR1 | UBD-3 | ubiquitin D | - | HPRD,BioGRID | 10200259 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | 24 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | 28 | 12 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 2 | 33 | 17 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MANN RESPONSE TO AMIFOSTINE DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH PROLIFERATION | 147 | 80 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN | 121 | 65 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| KAMMINGA EZH2 TARGETS | 41 | 26 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS UP | 33 | 18 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| KANG DOXORUBICIN RESISTANCE UP | 54 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESPONSE | 40 | 25 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D DN | 64 | 35 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |