Gene Page: MARK3
Summary ?
| GeneID | 4140 |
| Symbol | MARK3 |
| Synonyms | CTAK1|KP78|PAR1A|Par-1a |
| Description | microtubule affinity regulating kinase 3 |
| Reference | MIM:602678|HGNC:HGNC:6897|Ensembl:ENSG00000075413|HPRD:04058|Vega:OTTHUMG00000171789 |
| Gene type | protein-coding |
| Map location | 14q32.32 |
| Pascal p-value | 8.489E-7 |
| Sherlock p-value | 0.463 |
| Fetal beta | 0.385 |
| DMG | 1 (# studies) |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23090046 | 14 | 104094619 | MARK3 | 4.73E-5 | -2.428 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
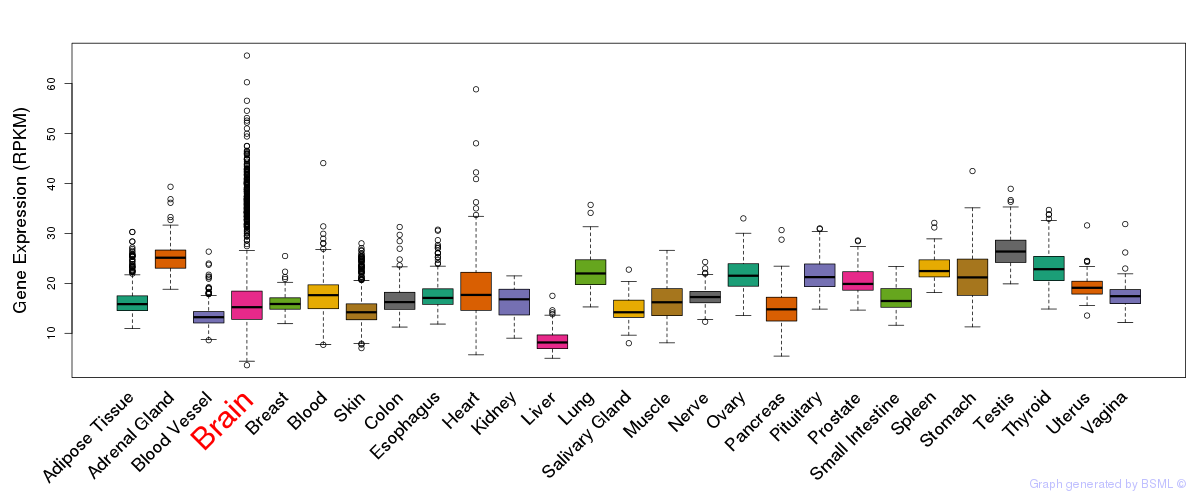
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP6V0C | ATP6C | ATP6L | ATPL | VATL | Vma3 | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | Two-hybrid | BioGRID | 16169070 |
| CHGB | SCG1 | chromogranin B (secretogranin 1) | Two-hybrid | BioGRID | 16169070 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| PMPCB | Beta-MPP | MPP11 | MPPB | MPPP52 | peptidase (mitochondrial processing) beta | Two-hybrid | BioGRID | 16169070 |
| PNPLA2 | 1110001C14Rik | ATGL | DESNUTRIN | DKFZp667M109 | FP17548 | PEDF-R | TTS-2.2 | TTS2 | patatin-like phospholipase domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| RASGRF1 | CDC25 | CDC25L | GNRP | GRF1 | GRF55 | H-GRF55 | PP13187 | Ras protein-specific guanine nucleotide-releasing factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9543386 |
| SAAL1 | FLJ41463 | serum amyloid A-like 1 | Two-hybrid | BioGRID | 16169070 |
| SFN | YWHAS | stratifin | Affinity Capture-MS Two-hybrid | BioGRID | 15778465 |16189514 |
| TCEA2 | TFIIS | transcription elongation factor A (SII), 2 | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| VPS11 | END1 | PEP5 | RNF108 | hVPS11 | vacuolar protein sorting 11 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| EHRLICH ICF SYNDROM UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |