Gene Page: MATK
Summary ?
| GeneID | 4145 |
| Symbol | MATK |
| Synonyms | CHK|CTK|HHYLTK|HYL|HYLTK|Lsk |
| Description | megakaryocyte-associated tyrosine kinase |
| Reference | MIM:600038|HGNC:HGNC:6906|Ensembl:ENSG00000007264|HPRD:02496|Vega:OTTHUMG00000180916 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.204 |
| Sherlock p-value | 0.005 |
| Fetal beta | -1.624 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12620556 | chr2 | 159196770 | MATK | 4145 | 0.12 | trans | ||
| rs6973230 | chr7 | 146072498 | MATK | 4145 | 0.15 | trans | ||
| rs717344 | chr16 | 5039247 | MATK | 4145 | 0.2 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
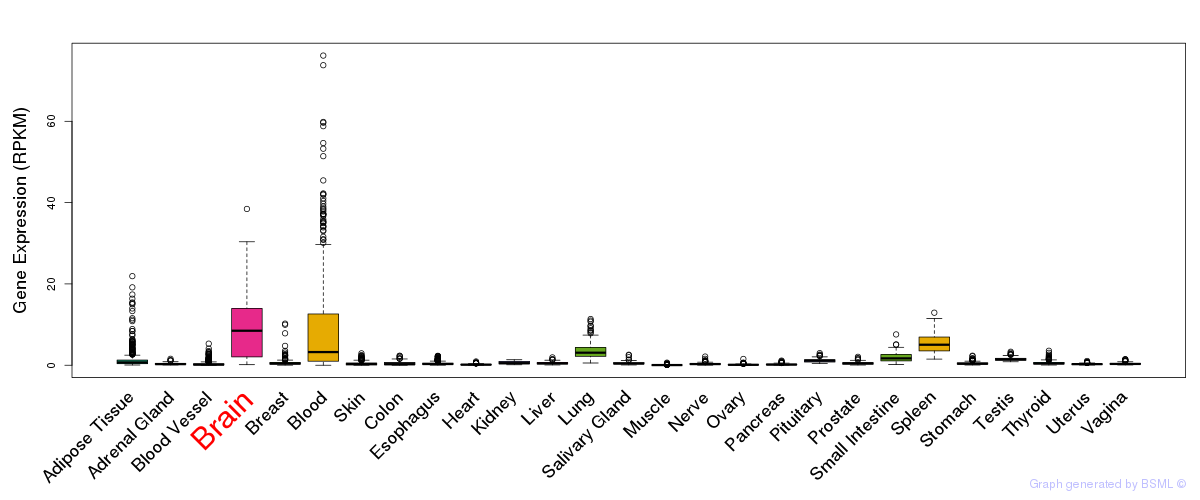
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMAD4 | 0.93 | 0.85 |
| GNA13 | 0.93 | 0.91 |
| PDCL | 0.93 | 0.84 |
| PPM1D | 0.92 | 0.87 |
| SMARCC1 | 0.92 | 0.85 |
| STAG1 | 0.91 | 0.88 |
| SFRS1 | 0.91 | 0.85 |
| TNPO1 | 0.91 | 0.86 |
| NONO | 0.91 | 0.82 |
| ALDH18A1 | 0.91 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.68 | -0.75 |
| S100B | -0.62 | -0.72 |
| IFI27 | -0.62 | -0.75 |
| AF347015.31 | -0.61 | -0.75 |
| FXYD1 | -0.61 | -0.74 |
| MT-CO2 | -0.61 | -0.76 |
| AF347015.27 | -0.61 | -0.73 |
| CA4 | -0.60 | -0.70 |
| HLA-F | -0.60 | -0.65 |
| AF347015.33 | -0.59 | -0.71 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN | 100 | 63 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE ALL DN | 11 | 8 | All SZGR 2.0 genes in this pathway |