Gene Page: MBP
Summary ?
| GeneID | 4155 |
| Symbol | MBP |
| Synonyms | - |
| Description | myelin basic protein |
| Reference | MIM:159430|HGNC:HGNC:6925|Ensembl:ENSG00000197971|HPRD:01158|Vega:OTTHUMG00000132874 |
| Gene type | protein-coding |
| Map location | 18q23 |
| Pascal p-value | 0.076 |
| DMG | 2 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_PocklingtonH1 G2Cdb.human_Synaptosome G2Cdb.humanNRC CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0328 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23470914 | 18 | 74800080 | MBP | 9.53E-6 | 0.661 | 0.013 | DMG:Wockner_2014 |
| cg20801056 | 18 | 74844767 | MBP | 3.004E-4 | -0.368 | 0.04 | DMG:Wockner_2014 |
| cg15407373 | 18 | 74800029 | MBP | 3.788E-4 | 0.363 | 0.043 | DMG:Wockner_2014 |
| cg18362936 | 18 | 74844011 | MBP | 6.87E-8 | -0.01 | 1.68E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1733504 | chr3 | 71372796 | MBP | 4155 | 0.03 | trans | ||
| rs1290499 | chr3 | 71393339 | MBP | 4155 | 0.08 | trans | ||
| rs6448932 | chr4 | 12595798 | MBP | 4155 | 0.17 | trans | ||
| rs17057381 | chr8 | 27416800 | MBP | 4155 | 0.15 | trans | ||
| rs758167 | chr12 | 1915558 | MBP | 4155 | 0.05 | trans |
Section II. Transcriptome annotation
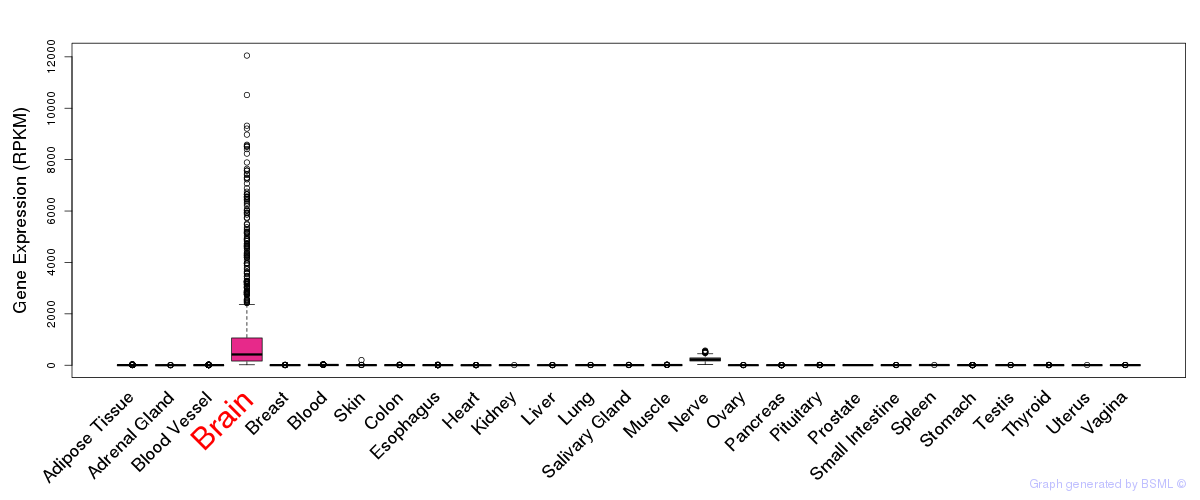
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019911 | structural constituent of myelin sheath | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 2434243 |
| GO:0008366 | axon ensheathment | TAS | neuron, axon (GO term level: 12) | 2434243 |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 2434243 |
| GO:0006955 | immune response | TAS | 7504278 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043209 | myelin sheath | IEA | neuron, axon, oligodendrocyte, Glial (GO term level: 7) | - |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BMP2K | BIKE | DKFZp434K0614 | DKFZp434P0116 | HRIHFB2017 | BMP2 inducible kinase | - | HPRD | 11500515 |
| CLK1 | CLK | CLK/STY | STY | CDC-like kinase 1 | - | HPRD | 8798720 |
| CRKRS | CRK7 | CRKR | KIAA0904 | Cdc2-related kinase, arginine/serine-rich | - | HPRD | 11683387 |
| CTDSP1 | NLIIF | SCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | - | HPRD | 14743429 |
| DYRK1B | MIRK | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | - | HPRD | 10910078 |
| EFHA1 | 1110008L20Rik | FLJ25016 | FLJ34588 | EF-hand domain family, member A1 | - | HPRD | 11124993 |
| HLA-DRA | HLA-DRA1 | major histocompatibility complex, class II, DR alpha | - | HPRD,BioGRID | 9782128 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD | 12237860 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | MBP interacts with MAPK1. This interaction was modeled on a demonstrated interaction between MBP from an unspecified species, and monkey and mouse MAPK1. | BIND | 9792705 |
| MAPK13 | MGC99536 | PRKM13 | SAPK4 | p38delta | mitogen-activated protein kinase 13 | - | HPRD | 9207191 |
| MAPK15 | ERK7 | ERK8 | mitogen-activated protein kinase 15 | - | HPRD | 11875070 |
| MAPKAPK5 | PRAK | mitogen-activated protein kinase-activated protein kinase 5 | - | HPRD | 9628874 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | - | HPRD | 9155018 |
| MMP7 | MMP-7 | MPSL1 | PUMP-1 | matrix metallopeptidase 7 (matrilysin, uterine) | - | HPRD | 8786845 |
| NEK9 | DKFZp434D0935 | MGC138306 | MGC16714 | NERCC | NERCC1 | Nek8 | NIMA (never in mitosis gene a)- related kinase 9 | - | HPRD | 11864968 |
| PLP1 | MMPL | PLP | PLP/DM20 | PMD | SPG2 | proteolipid protein 1 | - | HPRD,BioGRID | 2467009 |6083474 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | PKC-alpha phosphorylates and interacts with MBP. This interaction was modeled on a demonstrated interaction between PKC-alpha and MBP, both from unspecified species. | BIND | 10383403 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | PKC-delta phosphorylates and interacts with MBP. This interaction was modeled on a demonstrated interaction between PKC-delta and MBP, both from unspecified species. | BIND | 10383403 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | PKC-lambda phosphorylates and interacts with MBP. This interaction was modeled on a demonstrated interaction between PKC-lambda and MBP, both from unspecified species. | BIND | 10383403 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | PKC-zeta phosphorylates and interacts with MBP. This interaction was modeled on a demonstrated interaction between PKC-zeta and MBP, both from unspecified species. | BIND | 10383403 |
| PRMT5 | HRMT1L5 | IBP72 | JBP1 | SKB1 | SKB1Hs | protein arginine methyltransferase 5 | - | HPRD,BioGRID | 11152681 |
| ROCK1 | MGC131603 | MGC43611 | P160ROCK | PRO0435 | Rho-associated, coiled-coil containing protein kinase 1 | ROCK1 (ROCK) interacts with and phosphorylates MBP. | BIND | 15797222 |
| ROCK2 | KIAA0619 | Rho-associated, coiled-coil containing protein kinase 2 | ROCK2 (ROCK) interacts with and phosphorylates MBP. | BIND | 15797222 |
| RP6-213H19.1 | MASK | MST4 | serine/threonine protein kinase MST4 | - | HPRD | 11641781 |
| SRPK1 | SFRSK1 | SFRS protein kinase 1 | - | HPRD | 10390541 |
| STK16 | FLJ39635 | KRCT | MPSK | PKL12 | TSF1 | serine/threonine kinase 16 | - | HPRD | 10947953 |
| STK39 | DCHT | DKFZp686K05124 | PASK | SPAK | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | - | HPRD,BioGRID | 10980603 |
| STK4 | DKFZp686A2068 | KRS2 | MST1 | YSK3 | serine/threonine kinase 4 | - | HPRD | 7665586 |
| TLK1 | KIAA0137 | PKU-beta | tousled-like kinase 1 | - | HPRD | 10523312 |
| TLK2 | MGC44450 | PKU-ALPHA | tousled-like kinase 2 | - | HPRD | 10523312 |
| TRPM7 | CHAK | CHAK1 | FLJ20117 | FLJ25718 | LTRPC7 | TRP-PLIK | transient receptor potential cation channel, subfamily M, member 7 | - | HPRD,BioGRID | 14594813 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT UP | 21 | 19 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 1 UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS STIMULATED UP | 29 | 19 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO CYCLOPHOSPHAMIDE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| GOBERT CORE OLIGODENDROCYTE DIFFERENTIATION | 40 | 28 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 406 | 413 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-503 | 407 | 413 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.