| KEGG CITRATE CYCLE TCA CYCLE
| 32 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG PYRUVATE METABOLISM
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG GLYOXYLATE AND DICARBOXYLATE METABOLISM
| 16 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG PROXIMAL TUBULE BICARBONATE RECLAMATION
| 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KREB PATHWAY
| 8 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCONEOGENESIS
| 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM
| 69 | 42 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN
| 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHOI ATL CHRONIC VS ACUTE DN
| 18 | 10 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN
| 362 | 238 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN
| 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP
| 1142 | 669 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP
| 117 | 85 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS DN
| 37 | 25 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE DN
| 51 | 35 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN
| 712 | 443 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4
| 261 | 153 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP
| 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS
| 775 | 494 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN
| 52 | 34 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN
| 546 | 351 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN
| 64 | 42 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP
| 555 | 346 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR
| 532 | 309 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL
| 63 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN
| 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN
| 87 | 59 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN
| 123 | 76 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR
| 149 | 84 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA DN
| 29 | 15 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP
| 88 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE
| 77 | 50 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7
| 98 | 63 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP
| 388 | 234 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP
| 425 | 253 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN
| 79 | 45 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP
| 198 | 128 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN
| 102 | 65 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP
| 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP
| 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP
| 408 | 276 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES
| 389 | 245 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN
| 68 | 45 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS
| 535 | 325 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS
| 32 | 23 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP
| 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP
| 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL
| 129 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
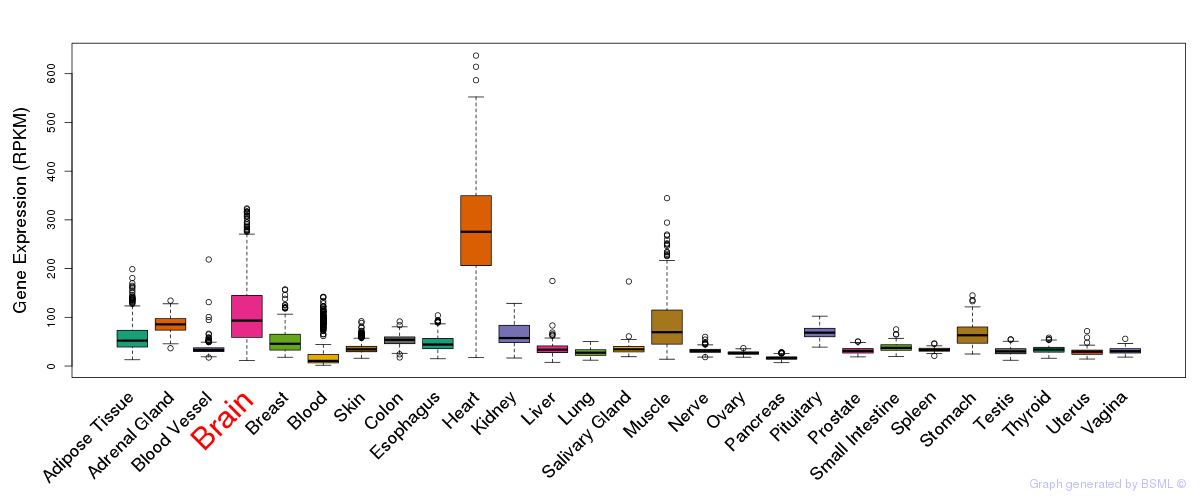
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation