Gene Page: MAP3K5
Summary ?
| GeneID | 4217 |
| Symbol | MAP3K5 |
| Synonyms | ASK1|MAPKKK5|MEKK5 |
| Description | mitogen-activated protein kinase kinase kinase 5 |
| Reference | MIM:602448|HGNC:HGNC:6857|Ensembl:ENSG00000197442|HPRD:03904|Vega:OTTHUMG00000015647 |
| Gene type | protein-coding |
| Map location | 6q22.33 |
| Pascal p-value | 0.383 |
| Sherlock p-value | 0.444 |
| Fetal beta | -2.716 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.139 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07868909 | 6 | 137114217 | MAP3K5 | 2.02E-9 | -0.038 | 1.64E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17034560 | chr1 | 10229157 | MAP3K5 | 4217 | 0.08 | trans |
Section II. Transcriptome annotation
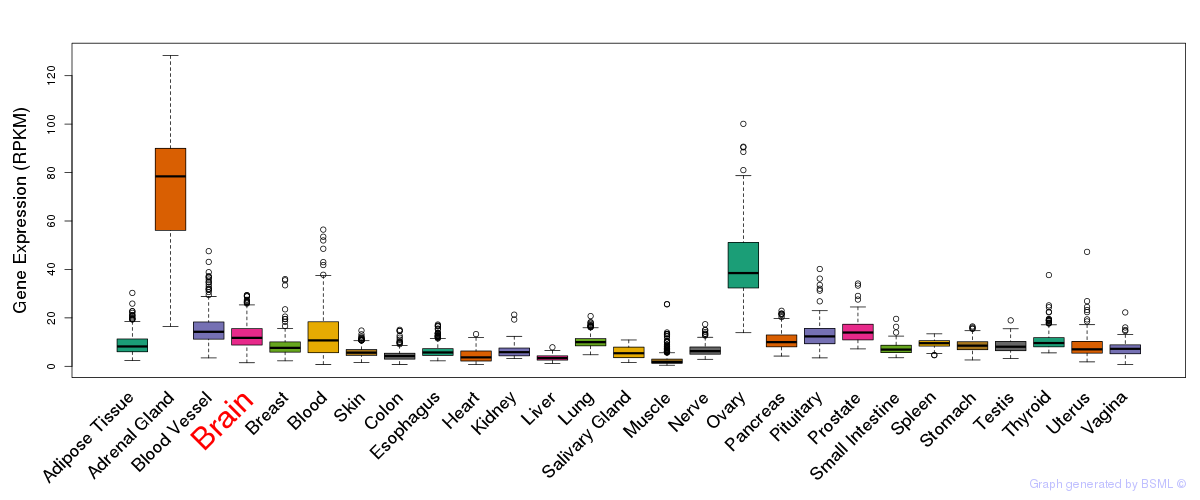
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GLI2 | 0.89 | 0.51 |
| KIFC1 | 0.87 | 0.49 |
| FOXM1 | 0.87 | 0.51 |
| IQGAP3 | 0.87 | 0.42 |
| NOTCH1 | 0.87 | 0.53 |
| CDT1 | 0.86 | 0.52 |
| PLK1 | 0.86 | 0.45 |
| CDC20 | 0.86 | 0.56 |
| KIF18B | 0.85 | 0.51 |
| MYBL2 | 0.85 | 0.48 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.27 | -0.36 |
| CCNI2 | -0.27 | -0.34 |
| RAPGEF4 | -0.26 | -0.35 |
| SERPINI1 | -0.26 | -0.33 |
| CKMT1A | -0.25 | -0.33 |
| NPM2 | -0.25 | -0.31 |
| CKMT1B | -0.25 | -0.27 |
| OSBPL1A | -0.25 | -0.33 |
| AF347015.27 | -0.25 | -0.33 |
| CHN1 | -0.24 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IDA | 17210579 | |
| GO:0005524 | ATP binding | IDA | 17210579 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004709 | MAP kinase kinase kinase activity | IDA | 17210579 | |
| GO:0008656 | caspase activator activity | IDA | 14761963 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | IDA | 11920685 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000165 | MAPKKK cascade | IDA | 17210579 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007257 | activation of JNK activity | TAS | 8974401 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0008624 | induction of apoptosis by extracellular signals | TAS | 8974401 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | Affinity Capture-Western | BioGRID | 11090355 |
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | - | HPRD | 11356842 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD,BioGRID | 11416155 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Affinity Capture-Western | BioGRID | 12820963 |
| DAB2IP | AF9Q34 | AIP1 | DIP1/2 | FLJ39072 | KIAA1743 | DAB2 interacting protein | - | HPRD | 12813029 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD,BioGRID | 9743501 |
| DUSP19 | DUSP17 | LMWDSP3 | MGC138210 | SKRP1 | TS-DSP1 | dual specificity phosphatase 19 | - | HPRD,BioGRID | 11959862 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 12473108 |
| ERN1 | FLJ30999 | IRE1 | IRE1P | MGC163277 | MGC163279 | endoplasmic reticulum to nucleus signaling 1 | - | HPRD,BioGRID | 12050113 |
| GADD45B | DKFZp566B133 | GADD45BETA | MYD118 | growth arrest and DNA-damage-inducible, beta | Affinity Capture-Western Reconstituted Complex | BioGRID | 14743220 |
| GLRX | GRX | GRX1 | MGC117407 | glutaredoxin (thioltransferase) | GLRX (GRX) interacts with MAP3K5 (ASK1). | BIND | 12244106 |
| GLRX | GRX | GRX1 | MGC117407 | glutaredoxin (thioltransferase) | - | HPRD,BioGRID | 12244106 |
| GSTM1 | GST1 | GSTM1-1 | GSTM1a-1a | GSTM1b-1b | GTH4 | GTM1 | H-B | MGC26563 | MU | MU-1 | glutathione S-transferase mu 1 | - | HPRD,BioGRID | 11278289 |12077134 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD,BioGRID | 12391142 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | Biochemical Activity Reconstituted Complex | BioGRID | 11689443 |12820963 |
| MAP2K6 | MAPKK6 | MEK6 | MKK6 | PRKMK6 | SAPKK3 | mitogen-activated protein kinase kinase 6 | MKK6 interacts with ASK1. This interaction was modeled on a demonstrated interaction between human MKK6 and ASK1 from an unspecified species. | BIND | 15866172 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | Affinity Capture-Western | BioGRID | 11959862 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | - | HPRD | 12912994 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD,BioGRID | 11920685 |
| MAP3K6 | ASK2 | MAPKKK6 | MGC125653 | MGC20114 | mitogen-activated protein kinase kinase kinase 6 | - | HPRD,BioGRID | 9875215 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD,BioGRID | 10921914 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD,BioGRID | 12189133 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | Affinity Capture-Western | BioGRID | 14607843 |
| PDCD6 | ALG-2 | FLJ46208 | MGC111017 | MGC119050 | MGC9123 | PEF1B | programmed cell death 6 | - | HPRD,BioGRID | 12372597 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD,BioGRID | 11689443 |
| QARS | GLNRS | PRO2195 | glutaminyl-tRNA synthetase | - | HPRD,BioGRID | 11096076 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 11427728 |
| RB1CC1 | CC1 | DRAGOU14 | FIP200 | RB1-inducible coiled-coil 1 | Affinity Capture-Western | BioGRID | 17015619 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | Affinity Capture-Western | BioGRID | 9774977 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 9774977 |10523862 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | Affinity Capture-Western | BioGRID | 9774977 |
| TRAF5 | MGC:39780 | RNF84 | TNF receptor-associated factor 5 | - | HPRD,BioGRID | 10523862 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 10523862 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9564042 |11689443 |12089063 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | TXN (TRX) interacts with MAP3K5 (ASK1). This interaction was modeled on a demonstrated interaction between human TXN and MAP3K5 from an unspecified species. | BIND | 12244106 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | - | HPRD | 9564042 |12089063 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 10411906|11336675 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA 41BB PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN | 56 | 39 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 1 | 46 | 33 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| ZHANG ADIPOGENESIS BY BMP7 | 14 | 12 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-145 | 368 | 374 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-17-5p/20/93.mr/106/519.d | 98 | 105 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-199 | 367 | 373 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-23 | 384 | 390 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-5p | 117 | 123 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 384 | 390 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-375 | 372 | 378 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.