Gene Page: MEN1
Summary ?
| GeneID | 4221 |
| Symbol | MEN1 |
| Synonyms | MEAI|SCG2 |
| Description | menin 1 |
| Reference | MIM:613733|HGNC:HGNC:7010|Ensembl:ENSG00000133895|HPRD:00564|Vega:OTTHUMG00000045366 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.099 |
| Sherlock p-value | 0.001 |
| Fetal beta | -0.212 |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
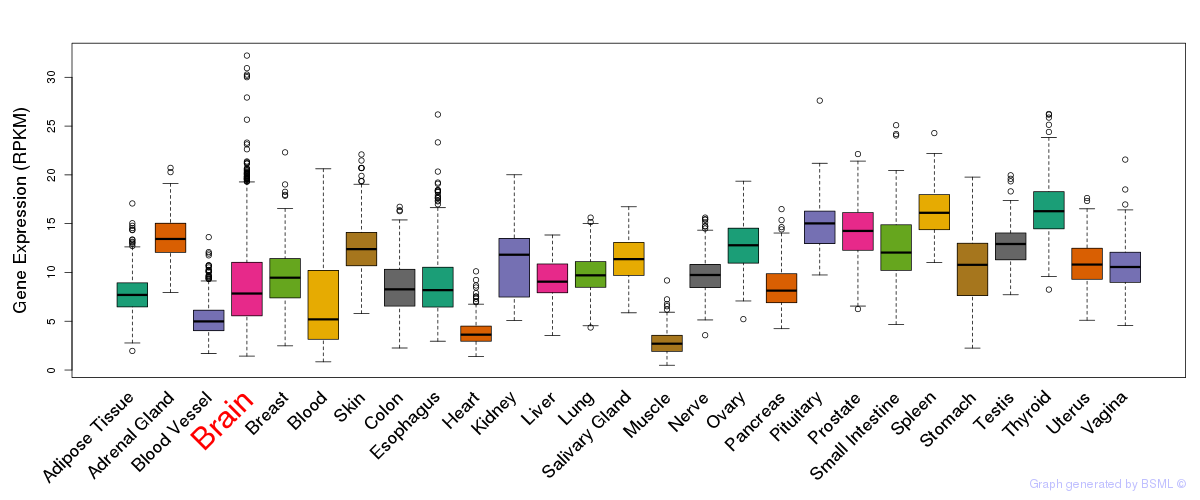
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASH2L | ASH2 | ASH2L1 | ASH2L2 | Bre2 | ash2 (absent, small, or homeotic)-like (Drosophila) | Reconstituted Complex | BioGRID | 15199122 |
| DBF4 | ASK | CHIF | DBF4A | ZDBF1 | DBF4 homolog (S. cerevisiae) | Menin interacts with ASK. | BIND | 15374998 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12874027 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | - | HPRD,BioGRID | 12169273 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Co-purification | BioGRID | 15199122 |
| HCFC2 | FLJ94012 | HCF-2 | HCF2 | host cell factor C2 | Co-purification | BioGRID | 15199122 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD,BioGRID | 9989505 |12226747 |
| JUND | AP-1 | jun D proto-oncogene | Menin directly represses JunD-activated transcription | BIND | 10500243 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 11526476 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | - | HPRD,BioGRID | 11526476 |
| RBBP5 | RBQ3 | SWD1 | retinoblastoma binding protein 5 | Co-purification | BioGRID | 15199122 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 11526476 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | - | HPRD,BioGRID | 12509449 |
| RPRM | FLJ90327 | REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate | Affinity Capture-MS | BioGRID | 17353931 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 11274402 |
| VIM | FLJ36605 | vimentin | menin interacts with vimentin | BIND | 12169273 |
| VIM | FLJ36605 | vimentin | - | HPRD,BioGRID | 12169273 |
| WDR5 | BIG-3 | SWD3 | WD repeat domain 5 | Co-purification | BioGRID | 15199122 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |