Gene Page: MAP3K10
Summary ?
| GeneID | 4294 |
| Symbol | MAP3K10 |
| Synonyms | MEKK10|MLK2|MST |
| Description | mitogen-activated protein kinase kinase kinase 10 |
| Reference | MIM:600137|HGNC:HGNC:6849|Ensembl:ENSG00000130758|HPRD:02533|Vega:OTTHUMG00000182591 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.484 |
| Sherlock p-value | 0.659 |
| Fetal beta | -0.627 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20005624 | 19 | 40697059 | MAP3K10 | 1.36E-9 | -0.015 | 1.37E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
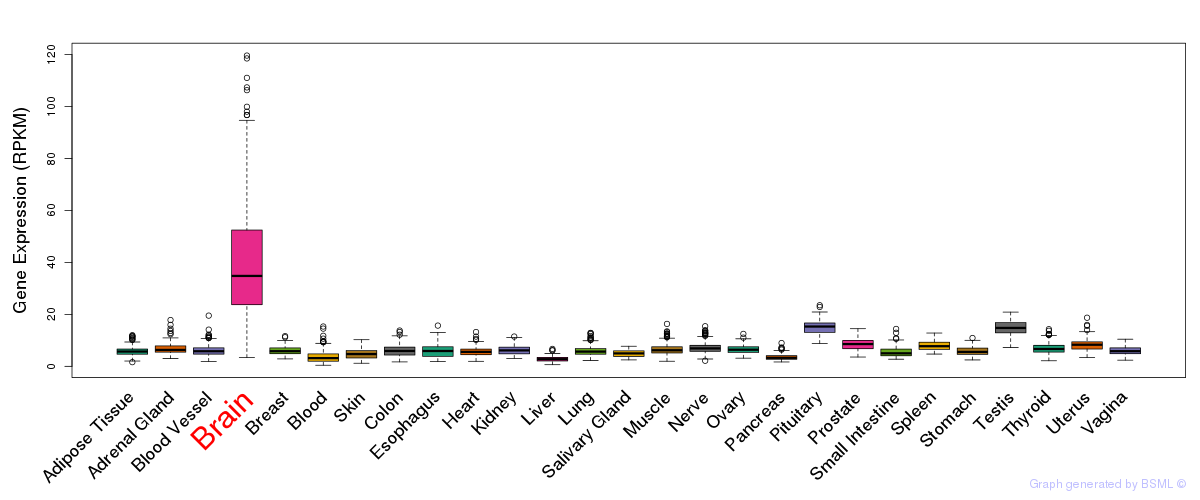
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AP000349.1 | 0.99 | 0.90 |
| GPC2 | 0.89 | 0.84 |
| SBK1 | 0.89 | 0.82 |
| ACTG1 | 0.88 | 0.79 |
| C6orf134 | 0.88 | 0.84 |
| MYD88 | 0.87 | 0.82 |
| SRC | 0.87 | 0.80 |
| TMEM44 | 0.87 | 0.80 |
| LRRC61 | 0.87 | 0.83 |
| IGLON5 | 0.87 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.63 | -0.74 |
| HLA-F | -0.62 | -0.71 |
| ALDOC | -0.61 | -0.68 |
| CLU | -0.61 | -0.68 |
| AIFM3 | -0.61 | -0.68 |
| FBXO2 | -0.61 | -0.65 |
| PTH1R | -0.60 | -0.64 |
| LDHD | -0.60 | -0.65 |
| RGN | -0.59 | -0.68 |
| AP003117.1 | -0.59 | -0.66 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD,BioGRID | 9427749 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD,BioGRID | 12105200 |
| CNKSR1 | CNK | CNK1 | KSR | connector enhancer of kinase suppressor of Ras 1 | CNKSR1 (hCNK1) interacts with MAP3K10 (MLK2). This interaction was modelled on a demonstrated interaction between human CNKSR1 and MAP3K10 from an unspecified species. | BIND | 15753034 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 11152698 |
| DNM1 | DNM | dynamin 1 | Reconstituted Complex | BioGRID | 9742220 |
| DNM1L | DLP1 | DRP1 | DVLP | DYMPLE | DYNIV-11 | FLJ41912 | HDYNIV | VPS1 | dynamin 1-like | - | HPRD | 9742220 |
| HPCA | BDR2 | hippocalcin | Affinity Capture-Western | BioGRID | 9427749 |
| HTT | HD | IT15 | huntingtin | MLK2 SH3 domain interacts with Huntingtin | BIND | 10801775 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 10801775 |
| KIF17 | KIAA1405 | KIF17B | KIF3X | kinesin family member 17 | Two-hybrid | BioGRID | 9427749 |
| KIF3A | - | kinesin family member 3A | - | HPRD,BioGRID | 9427749 |
| KIF3B | HH0048 | KIAA0359 | kinesin family member 3B | - | HPRD | 9427749 |
| KIFAP3 | FLJ22818 | KAP3 | SMAP | Smg-GDS | dJ190I16.1 | kinesin-associated protein 3 | - | HPRD,BioGRID | 9427749 |
| KRTAP3P1 | KAP3A | keratin associated protein 3 pseudogene 1 | Affinity Capture-Western | BioGRID | 9427749 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD,BioGRID | 9182538 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| MAPK8IP2 | IB2 | JIP2 | PRKM8IPL | mitogen-activated protein kinase 8 interacting protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10490659 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | Affinity Capture-Western Biochemical Activity Two-hybrid | BioGRID | 12881483 |
| PHB | PHB1 | prohibitin | - | HPRD | 9629920 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Reconstituted Complex | BioGRID | 9427749 |
| RACGAP1 | HsCYK-4 | ID-GAP | MgcRacGAP | Rac GTPase activating protein 1 | - | HPRD | 9427749 |
| SH3RF1 | FLJ21602 | KIAA1494 | POSH | RNF142 | SH3MD2 | SH3 domain containing ring finger 1 | - | HPRD | 14504284 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD | 9629920 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 9427749 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| KUUSELO PANCREATIC CANCER 19Q13 AMPLIFICATION | 35 | 22 | All SZGR 2.0 genes in this pathway |
| LIU IL13 MEMORY MODEL UP | 17 | 12 | All SZGR 2.0 genes in this pathway |