Gene Page: AFF1
Summary ?
| GeneID | 4299 |
| Symbol | AFF1 |
| Synonyms | AF4|MLLT2|PBM1 |
| Description | AF4/FMR2 family member 1 |
| Reference | MIM:159557|HGNC:HGNC:7135|Ensembl:ENSG00000172493|HPRD:08871|Vega:OTTHUMG00000130603 |
| Gene type | protein-coding |
| Map location | 4q21 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 0.535 |
| Fetal beta | -0.875 |
| DMG | 3 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11468363 | 4 | 87857418 | AFF1 | 8.55E-5 | 0.401 | 0.026 | DMG:Wockner_2014 |
| cg21046364 | 4 | 87871098 | AFF1 | 2.106E-4 | 0.293 | 0.035 | DMG:Wockner_2014 |
| cg16090392 | 4 | 87928170 | AFF1 | -0.032 | 0.58 | DMG:Nishioka_2013 | |
| cg18044720 | 4 | 87857031 | AFF1 | 3.4E-8 | -0.016 | 1.01E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
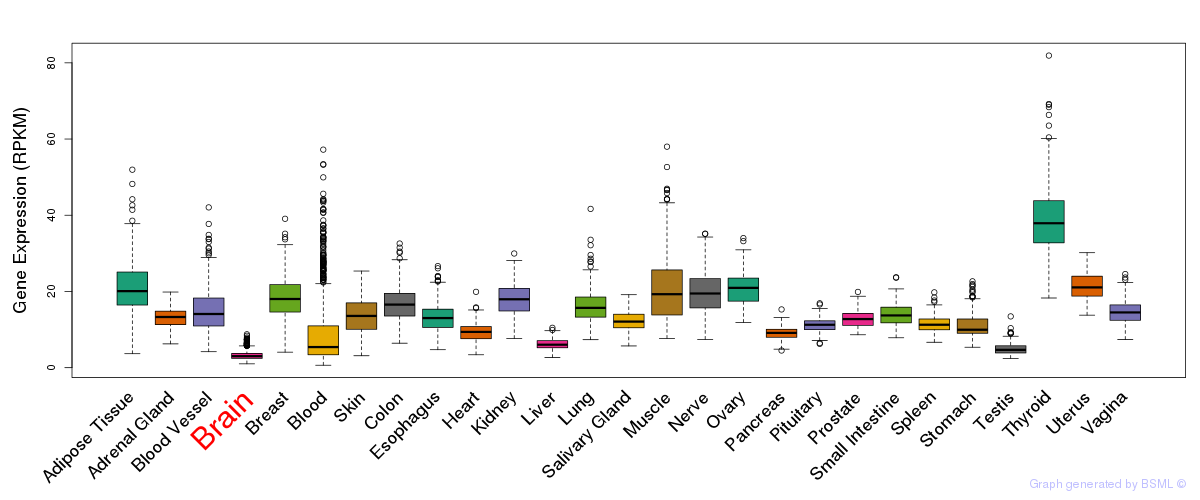
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRIM33 | 0.92 | 0.93 |
| MED13 | 0.91 | 0.94 |
| PDE4D | 0.90 | 0.92 |
| AFF4 | 0.90 | 0.95 |
| KIAA2022 | 0.90 | 0.94 |
| GSK3B | 0.90 | 0.93 |
| LASS6 | 0.90 | 0.91 |
| JMY | 0.90 | 0.92 |
| CHML | 0.90 | 0.93 |
| KLHDC10 | 0.90 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.64 | -0.80 |
| HSD17B14 | -0.64 | -0.74 |
| SERPINB6 | -0.64 | -0.70 |
| AIFM3 | -0.63 | -0.69 |
| AF347015.31 | -0.62 | -0.78 |
| HIGD1B | -0.62 | -0.80 |
| TSC22D4 | -0.62 | -0.71 |
| MT-CO2 | -0.62 | -0.79 |
| AC018755.7 | -0.61 | -0.68 |
| S100B | -0.61 | -0.74 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| HUMMERICH BENIGN SKIN TUMOR DN | 18 | 7 | All SZGR 2.0 genes in this pathway |
| HUMMERICH MALIGNANT SKIN TUMOR DN | 18 | 7 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HBZ | 41 | 27 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |