Gene Page: MYO6
Summary ?
| GeneID | 4646 |
| Symbol | MYO6 |
| Synonyms | DFNA22|DFNB37 |
| Description | myosin VI |
| Reference | MIM:600970|HGNC:HGNC:7605|Ensembl:ENSG00000196586|HPRD:02985|Vega:OTTHUMG00000015061 |
| Gene type | protein-coding |
| Map location | 6q13 |
| Pascal p-value | 0.748 |
| Sherlock p-value | 0.833 |
| Fetal beta | -0.049 |
| eGene | Cerebellar Hemisphere Cerebellum Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0086 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
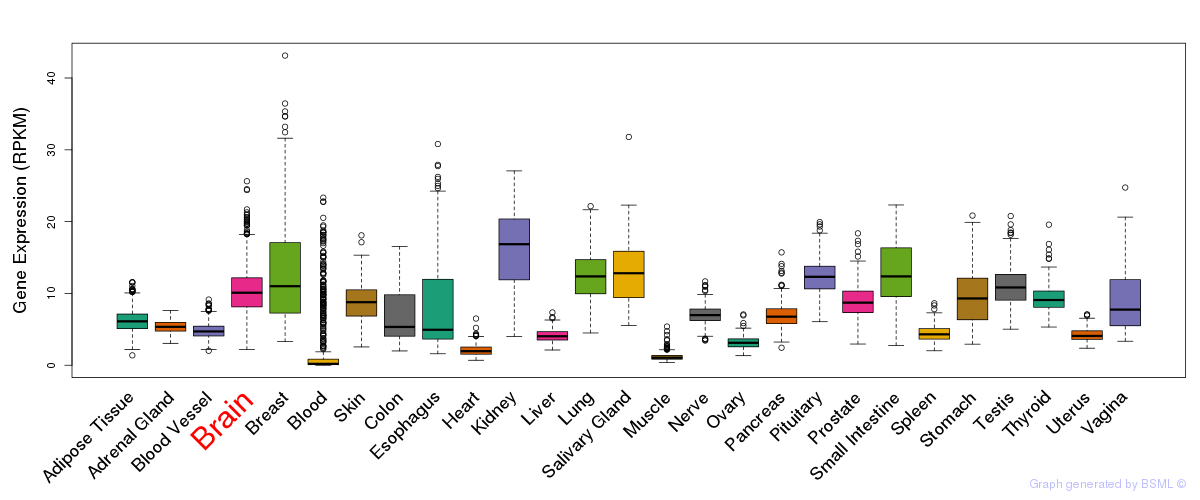
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003774 | motor activity | IEA | - | |
| GO:0003774 | motor activity | ISS | - | |
| GO:0005516 | calmodulin binding | ISS | - | |
| GO:0005516 | calmodulin binding | TAS | 12857860 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0051015 | actin filament binding | IDA | 9852149 | |
| GO:0051015 | actin filament binding | ISS | - | |
| GO:0042803 | protein homodimerization activity | NAS | 11728438 | |
| GO:0043531 | ADP binding | ISS | - | |
| GO:0060001 | minus-end directed microfilament motor activity | NAS | 10519557 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| GO:0007605 | sensory perception of sound | IDA | 11468689 | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0030048 | actin filament-based movement | ISS | - | |
| GO:0030048 | actin filament-based movement | NAS | 10519557 | |
| GO:0006897 | endocytosis | IMP | 15247260 | |
| GO:0006897 | endocytosis | ISS | - | |
| GO:0006886 | intracellular protein transport | ISS | - | |
| GO:0051046 | regulation of secretion | IMP | 15837803 | |
| GO:0030330 | DNA damage response, signal transduction by p53 class mediator | IDA | 16507995 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IMP | 16949370 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | IDA | 9852149 |16507995 | |
| GO:0005794 | Golgi apparatus | IDA | 16507995 | |
| GO:0005634 | nucleus | IDA | 16507995 | |
| GO:0005654 | nucleoplasm | IDA | 16948370 | |
| GO:0005737 | cytoplasm | IDA | 9852149 |16507995 |16949370 | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0005938 | cell cortex | ISS | - | |
| GO:0005905 | coated pit | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 9852149 | |
| GO:0048471 | perinuclear region of cytoplasm | ISS | - | |
| GO:0016461 | unconventional myosin complex | NAS | 8022818 | |
| GO:0016461 | unconventional myosin complex | TAS | 9259267 | |
| GO:0016591 | DNA-directed RNA polymerase II, holoenzyme | IDA | 16949370 | |
| GO:0031965 | nuclear membrane | IDA | 16507995 | |
| GO:0031941 | filamentous actin | IDA | 9852149 | |
| GO:0031941 | filamentous actin | ISS | - | |
| GO:0031941 | filamentous actin | ISS | - | |
| GO:0045334 | clathrin-coated endocytic vesicle | IDA | 11447109 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION DEGRADATION | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION TRAFFICKING | 27 | 19 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 8Q24 UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP | 40 | 27 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 UP | 57 | 34 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| REICHERT MITOSIS LIN9 TARGETS | 28 | 17 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-143 | 840 | 847 | 1A,m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-145 | 4484 | 4490 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-146 | 2840 | 2846 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-23 | 1478 | 1484 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-299-5p | 2265 | 2271 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.