Gene Page: NAP1L2
Summary ?
| GeneID | 4674 |
| Symbol | NAP1L2 |
| Synonyms | BPX |
| Description | nucleosome assembly protein 1 like 2 |
| Reference | MIM:300026|HGNC:HGNC:7638|Ensembl:ENSG00000186462|HPRD:02066|Vega:OTTHUMG00000021827 |
| Gene type | protein-coding |
| Map location | Xq13 |
| Sherlock p-value | 0.96 |
| TADA p-value | 9.98E-4 |
| Fetal beta | -2.504 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NAP1L2 | chrX | 72434103..72434104 | CT | C | NM_021963 | . | frameshift | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | NAP1L2 | 4674 | 0.17 | trans |
Section II. Transcriptome annotation
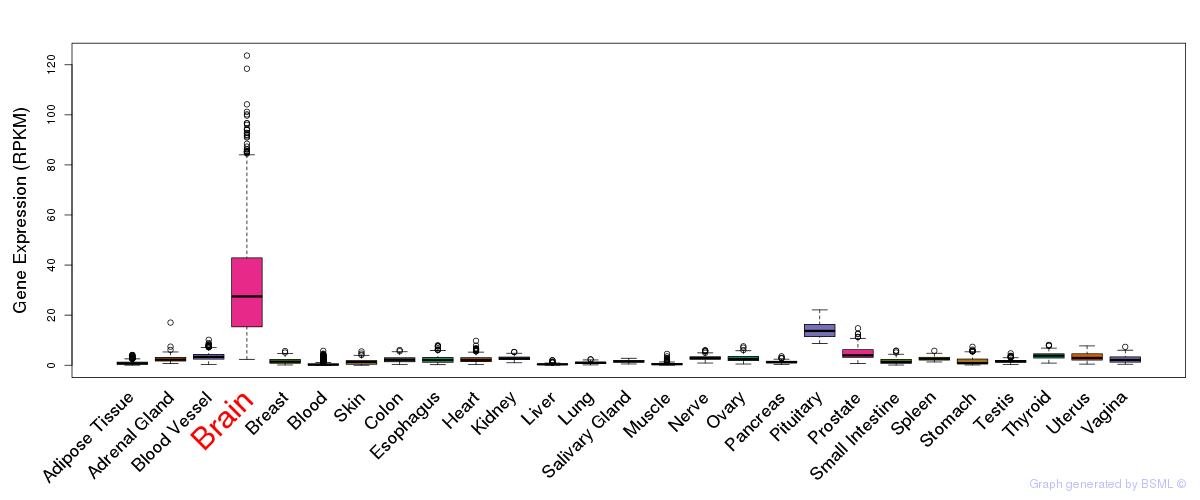
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TOMM5 | 0.89 | 0.87 |
| CHCHD1 | 0.86 | 0.86 |
| TXNDC17 | 0.86 | 0.85 |
| UQCRH | 0.85 | 0.82 |
| POLR2F | 0.84 | 0.83 |
| SNRPD2 | 0.84 | 0.83 |
| NDUFS4 | 0.84 | 0.83 |
| NDUFA12 | 0.82 | 0.82 |
| MRPS21 | 0.82 | 0.82 |
| MRPL27 | 0.82 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PEAR1 | -0.49 | -0.55 |
| TENC1 | -0.48 | -0.54 |
| LRRC32 | -0.46 | -0.56 |
| TNS1 | -0.45 | -0.53 |
| SEMA3G | -0.44 | -0.50 |
| FAM38A | -0.44 | -0.52 |
| EPAS1 | -0.44 | -0.52 |
| AHNAK | -0.44 | -0.54 |
| NPR1 | -0.43 | -0.48 |
| VWF | -0.43 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PYEON HPV POSITIVE TUMORS DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |