| LIU PROSTATE CANCER DN
| 481 | 290 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN
| 90 | 65 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN
| 175 | 82 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP
| 82 | 51 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP
| 450 | 256 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES UP
| 72 | 38 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP
| 112 | 71 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN
| 22 | 14 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN
| 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN
| 142 | 90 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN
| 143 | 83 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP
| 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN
| 781 | 465 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION UP
| 52 | 32 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER
| 178 | 108 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN
| 114 | 58 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN
| 193 | 112 | All SZGR 2.0 genes in this pathway |
| MURATA VIRULENCE OF H PILORI
| 24 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX
| 795 | 478 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT
| 108 | 73 | All SZGR 2.0 genes in this pathway |
| JI METASTASIS REPRESSED BY STK11
| 27 | 17 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN
| 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN
| 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN
| 59 | 32 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS DN
| 66 | 39 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX DN
| 21 | 11 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS
| 318 | 215 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP
| 412 | 249 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN
| 246 | 180 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP
| 71 | 48 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN
| 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3
| 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN
| 70 | 44 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP
| 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP
| 147 | 83 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN
| 701 | 446 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN
| 51 | 29 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN
| 23 | 15 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING UP
| 41 | 24 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP
| 288 | 168 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN
| 505 | 328 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1
| 467 | 251 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE
| 69 | 31 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
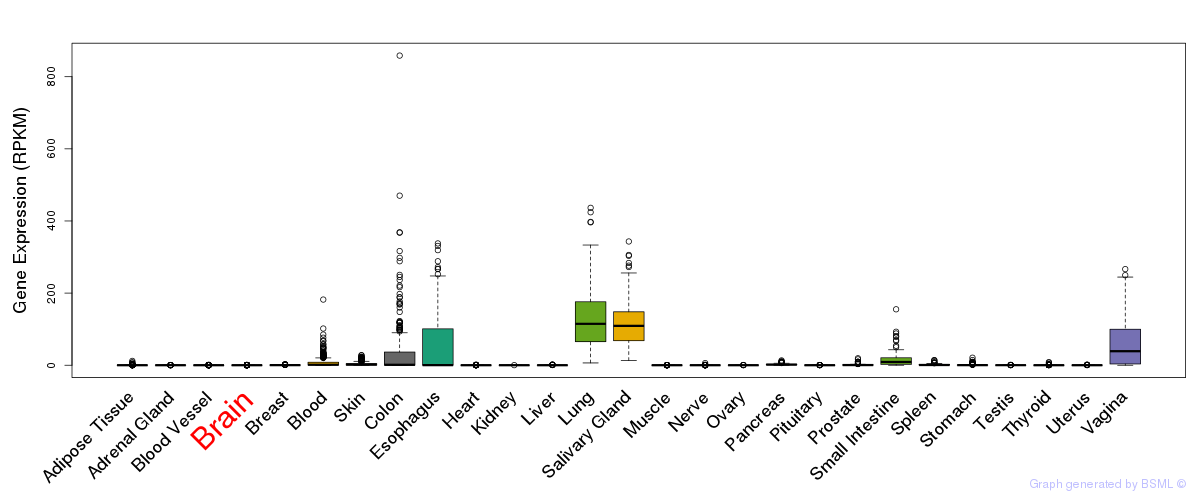
 eQTL annotation
eQTL annotation