| LIU SOX4 TARGETS DN
| 309 | 191 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP
| 115 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN
| 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN
| 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP
| 197 | 135 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION
| 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3
| 48 | 31 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5
| 147 | 89 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP
| 390 | 236 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN
| 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN
| 428 | 306 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX1 UP
| 15 | 10 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN
| 45 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS
| 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS
| 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3
| 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS
| 652 | 441 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP
| 195 | 138 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN
| 517 | 309 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES
| 147 | 87 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP
| 19 | 14 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP
| 42 | 24 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION
| 405 | 264 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP
| 69 | 41 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE
| 35 | 28 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL
| 254 | 164 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP
| 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP
| 487 | 303 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP
| 428 | 266 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 11
| 57 | 40 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN
| 48 | 33 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER
| 22 | 19 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN
| 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP
| 49 | 32 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP
| 89 | 62 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION
| 368 | 247 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED
| 351 | 238 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3
| 102 | 76 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT
| 567 | 365 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP
| 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP
| 140 | 81 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP
| 134 | 85 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN
| 553 | 343 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN
| 244 | 157 | All SZGR 2.0 genes in this pathway |
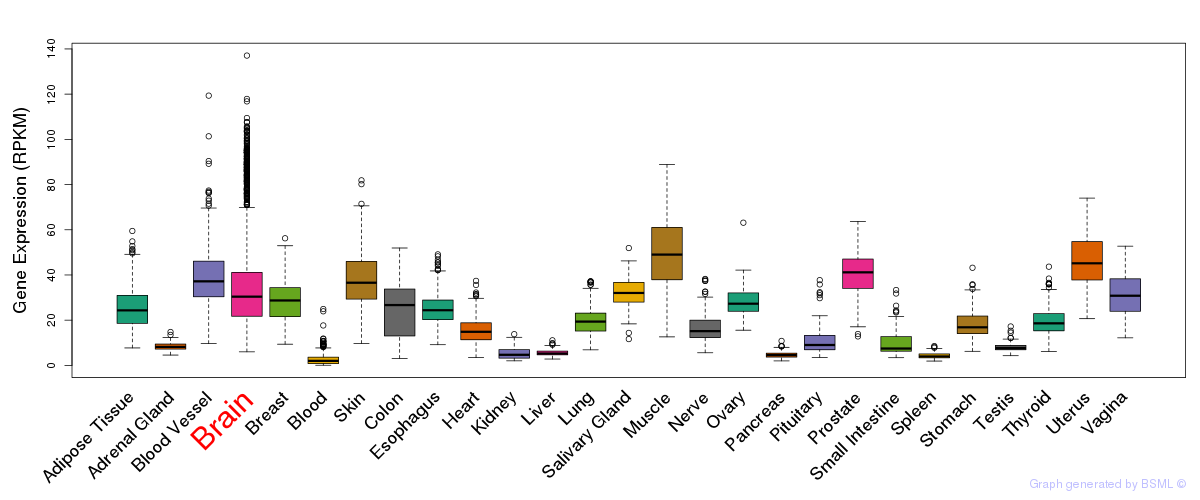
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation