Gene Page: NFKB1
Summary ?
| GeneID | 4790 |
| Symbol | NFKB1 |
| Synonyms | CVID12|EBP-1|KBF1|NF-kB1|NF-kappa-B|NF-kappaB|NFKB-p105|NFKB-p50|NFkappaB|p105|p50 |
| Description | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| Reference | MIM:164011|HGNC:HGNC:7794|Ensembl:ENSG00000109320|HPRD:01238|Vega:OTTHUMG00000161080 |
| Gene type | protein-coding |
| Map location | 4q24 |
| Pascal p-value | 0.113 |
| Sherlock p-value | 0.112 |
| Fetal beta | -0.303 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0798 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
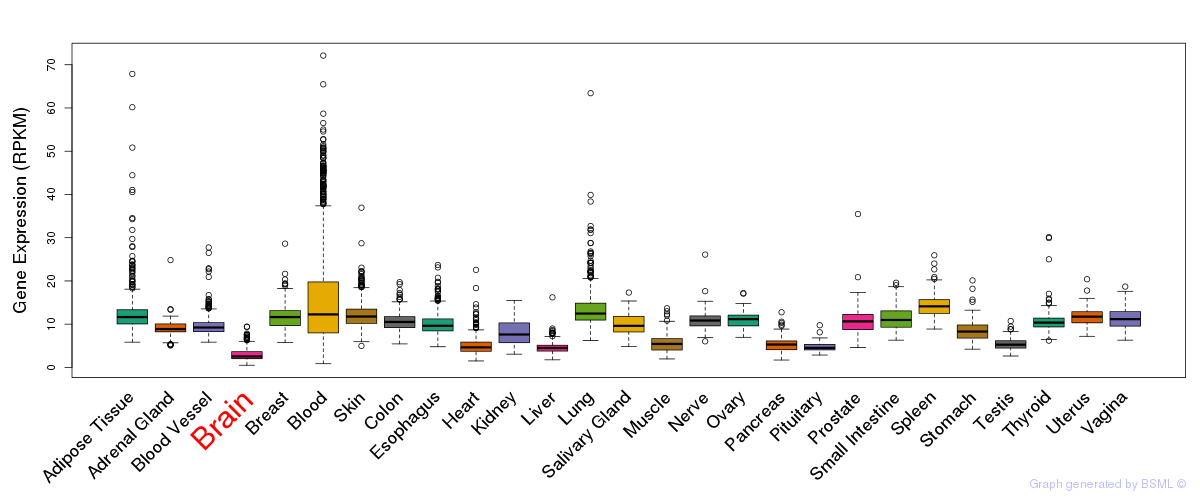
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OLIG1 | 0.82 | 0.89 |
| OLIG2 | 0.81 | 0.86 |
| SOX10 | 0.75 | 0.87 |
| HEPACAM | 0.71 | 0.81 |
| ZCCHC24 | 0.71 | 0.81 |
| NFATC1 | 0.70 | 0.67 |
| CHAD | 0.70 | 0.77 |
| SLC12A4 | 0.69 | 0.63 |
| PRR5L | 0.69 | 0.65 |
| CD82 | 0.68 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STMN1 | -0.45 | -0.56 |
| STMN2 | -0.44 | -0.55 |
| MAPK8 | -0.44 | -0.52 |
| POLB | -0.44 | -0.59 |
| ZNF551 | -0.44 | -0.52 |
| ZNF286 | -0.44 | -0.49 |
| PPP3CC | -0.43 | -0.54 |
| AC004017.1 | -0.43 | -0.50 |
| MED19 | -0.43 | -0.56 |
| FRG1 | -0.43 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 10928981 | |
| GO:0005515 | protein binding | IPI | 8196632 |11526476 |14624448 |16108830 |16306601 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 1992489 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0006954 | inflammatory response | TAS | 1992489 | |
| GO:0006916 | anti-apoptosis | TAS | 10811897 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0045941 | positive regulation of transcription | NAS | 8096091 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10723127 | |
| GO:0005634 | nucleus | IDA | 11819787 | |
| GO:0005654 | nucleoplasm | EXP | 10723127 | |
| GO:0005737 | cytoplasm | IDA | 11819787 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD | 8798655 |
| ATF3 | - | activating transcription factor 3 | - | HPRD | 7692236 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | p50 interacts with BCL-3. This interaction was modelled on a demonstrated interaction between p50 from an unspecified species and human BCL-3. | BIND | 15469820 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD | 10362352 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | Affinity Capture-Western Co-localization Co-purification Reconstituted Complex | BioGRID | 8428580 |10469655 |14678988 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | NFKB1 (p50) interacts with BCL3. | BIND | 15829968 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | The cIAP-2 promoter interacts with p50. | BIND | 15494311 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 12700228 |
| BTRC | BETA-TRCP | FBW1A | FBXW1 | FBXW1A | FWD1 | MGC4643 | bTrCP | bTrCP1 | betaTrCP | beta-transducin repeat containing | Affinity Capture-Western | BioGRID | 11158290 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | NFKB1 (p50) interacts with the CD82 (KAI1) promoter. | BIND | 15829968 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | - | HPRD | 12173051 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 1518839 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 10469655 |
| COPB2 | beta'-COP | coatomer protein complex, subunit beta 2 (beta prime) | Affinity Capture-MS | BioGRID | 14743216 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | NFKB1 (p50) interacts with CTNNB1 (beta-catenin). | BIND | 15829968 |
| DSP | DPI | DPII | desmoplakin | - | HPRD | 14743216 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Reconstituted Complex | BioGRID | 9368006 |
| ELF1 | - | E74-like factor 1 (ets domain transcription factor) | - | HPRD,BioGRID | 7862168 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | - | HPRD,BioGRID | 11036073 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD | 9094628 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD | 9468519 |
| G3BP2 | - | GTPase activating protein (SH3 domain) binding protein 2 | - | HPRD | 10969074 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD,BioGRID | 11425860 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 11931769 |
| HMGA2 | BABL | HMGI-C | HMGIC | LIPO | STQTL9 | high mobility group AT-hook 2 | - | HPRD,BioGRID | 12693954 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 12604365 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | - | HPRD | 14743216 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD | 10469655|11158290 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | Affinity Capture-Western Biochemical Activity | BioGRID | 10469655 |11158290 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | Affinity Capture-MS | BioGRID | 14743216 |
| IL2RA | CD25 | IDDM10 | IL2R | TCGFR | interleukin 2 receptor, alpha | in vitro in vivo | BioGRID | 2497520 |
| IL8 | CXCL8 | GCP-1 | GCP1 | LECT | LUCT | LYNAP | MDNCF | MONAP | NAF | NAP-1 | NAP1 | interleukin 8 | The IL-8 promoter interacts with p50. | BIND | 15494311 |
| IRF1 | IRF-1 | MAR | interferon regulatory factor 1 | - | HPRD | 8746784 |
| IRF2 | DKFZp686F0244 | IRF-2 | interferon regulatory factor 2 | - | HPRD,BioGRID | 8550813 |
| ITGB3BP | CENP-R | CENPR | HSU37139 | NRIF3 | TAP20 | integrin beta 3 binding protein (beta3-endonexin) | - | HPRD,BioGRID | 12244126 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | HTATIP (Tip60) interacts with NFKB1 (p50). | BIND | 15829968 |
| KLF5 | BTEB2 | CKLF | IKLF | Kruppel-like factor 5 (intestinal) | - | HPRD,BioGRID | 14573617 |
| KPNA3 | IPOA4 | SRP1gamma | SRP4 | hSRP1 | karyopherin alpha 3 (importin alpha 4) | Affinity Capture-MS | BioGRID | 14743216 |
| LYL1 | bHLHa18 | lymphoblastic leukemia derived sequence 1 | in vitro in vivo Two-hybrid | BioGRID | 10023675 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12150997 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | - | HPRD,BioGRID | 9950430 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | - | HPRD,BioGRID | 11526476 |
| MRCL3 | MLCB | MRLC3 | myosin regulatory light chain MRCL3 | - | HPRD | 14743216 |
| MTPN | FLJ31098 | FLJ99857 | GCDP | V-1 | myotrophin | - | HPRD,BioGRID | 11971907 |
| MYL6 | ESMLC | LC17-GI | LC17-NM | LC17A | LC17B | MLC1SM | MLC3NM | MLC3SM | myosin, light chain 6, alkali, smooth muscle and non-muscle | - | HPRD | 14743216 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | - | HPRD | 12618429 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | SRC-1 interacts with p50. This interaction is modelled on a demonstrated interaction between human SRC-1 and p50 from an unspecified species. | BIND | 9556555 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9556555 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex | BioGRID | 10847592 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Affinity Capture-Western | BioGRID | 12150997 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 10777532 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | Affinity Capture-MS Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 8428580 |14743216 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | NFKB1 (p50) forms a homodimer. | BIND | 15735750 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | p50 interacts with p50. This interaction was modelled on a demonstrated interaction between p50 and p50 both from an unspecified species. | BIND | 9556555 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | Affinity Capture-MS | BioGRID | 14743216 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD,BioGRID | 9738011 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | p50 associates with I-kappa-B-alpha. | BIND | 9135156 |
| NFKBIE | IKBE | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | - | HPRD | 9223487 |
| NFKBIE | IKBE | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9315679 |
| NFKBIZ | FLJ30225 | FLJ34463 | IKBZ | INAP | MAIL | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | Affinity Capture-Western | BioGRID | 11356851 |
| NKRF | ITBA4 | NRF | NFKB repressing factor | Reconstituted Complex | BioGRID | 10562553 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | - | HPRD,BioGRID | 8642313 |11418662 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 7823959 |8290595 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD | 11590148 |
| PPP4C | PP4 | PPH3 | PPX | protein phosphatase 4 (formerly X), catalytic subunit | - | HPRD,BioGRID | 9837938 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 14743216 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-MS Reconstituted Complex | BioGRID | 8798655 |14743216 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | NFKB1 (p50) interacts with RELA (p65). | BIND | 15735750 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | p50 interacts with p65. This interaction was modeled on a demonstrated interaction between mouse p50 and mouse p65. | BIND | 9738011 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD | 8649779 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | Affinity Capture-MS | BioGRID | 14743216 |
| RSF1 | HBXAP | RSF-1 | XAP8 | p325 | remodeling and spacing factor 1 | - | HPRD,BioGRID | 15242768 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | - | HPRD | 14743216 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | NFKB1 (p50) interacts with RUVBL2 (reptin). | BIND | 15829968 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 10075655 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 7933095 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | - | HPRD,BioGRID | 14743216 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | NFKB1 (NF-kB1/p105) interacts with an unspecified isoform of SPAG9 (JLP). | BIND | 14743216 |
| SPI1 | OF | PU.1 | SFPI1 | SPI-1 | SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | - | HPRD | 9094628 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12057007 |
| STAT6 | D12S1644 | IL-4-STAT | STAT6B | STAT6C | signal transducer and activator of transcription 6, interleukin-4 induced | - | HPRD,BioGRID | 9584180 |
| TBK1 | FLJ11330 | NAK | T2K | TANK-binding kinase 1 | Affinity Capture-MS | BioGRID | 14743216 |
| TNFSF11 | CD254 | ODF | OPGL | OPTB2 | RANKL | TRANCE | hRANKL2 | sOdf | tumor necrosis factor (ligand) superfamily, member 11 | - | HPRD | 10635328 |
| TNIP2 | ABIN-2 | ABIN2 | FLIP1 | KLIP | MGC4289 | TNFAIP3 interacting protein 2 | - | HPRD,BioGRID | 14743216 |15169888 |
| TNIP3 | ABIN-3 | FLJ21162 | LIND | TNFAIP3 interacting protein 3 | Affinity Capture-MS | BioGRID | 14743216 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western | BioGRID | 10646849 |
| TPR | - | translocated promoter region (to activated MET oncogene) | - | HPRD | 14743216 |
| TRIP4 | HsT17391 | thyroid hormone receptor interactor 4 | - | HPRD | 12077347 |
| TSC22D3 | DIP | DKFZp313A1123 | DSIPI | GILZ | TSC-22R | hDIP | TSC22 domain family, member 3 | - | HPRD,BioGRID | 11468175 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | - | HPRD | 14743216 |
| TUBB6 | HsT1601 | MGC132410 | MGC4083 | TUBB-5 | tubulin, beta 6 | - | HPRD | 14743216 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | - | HPRD,BioGRID | 7788295 |
| UBE2K | DKFZp564C1216 | DKFZp686J24237 | E2-25K | HIP2 | HYPG | LIG | ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | - | HPRD | 9535861 |
| UNC5CL | MGC34763 | ZUD | unc-5 homolog C (C. elegans)-like | - | HPRD,BioGRID | 14769797 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-30-3p | 562 | 568 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-323 | 480 | 486 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-9 | 29 | 35 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.