Gene Page: NFYB
Summary ?
| GeneID | 4801 |
| Symbol | NFYB |
| Synonyms | CBF-A|CBF-B|HAP3|NF-YB |
| Description | nuclear transcription factor Y subunit beta |
| Reference | MIM:189904|HGNC:HGNC:7805|Ensembl:ENSG00000120837|HPRD:01794|Vega:OTTHUMG00000170176 |
| Gene type | protein-coding |
| Map location | 12q23.3 |
| Pascal p-value | 0.151 |
| Fetal beta | 0.977 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12062545 | 12 | 104532339 | NFYB | 2.56E-9 | -0.01 | 1.88E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16836102 | chr4 | 4716366 | NFYB | 4801 | 0.09 | trans |
Section II. Transcriptome annotation
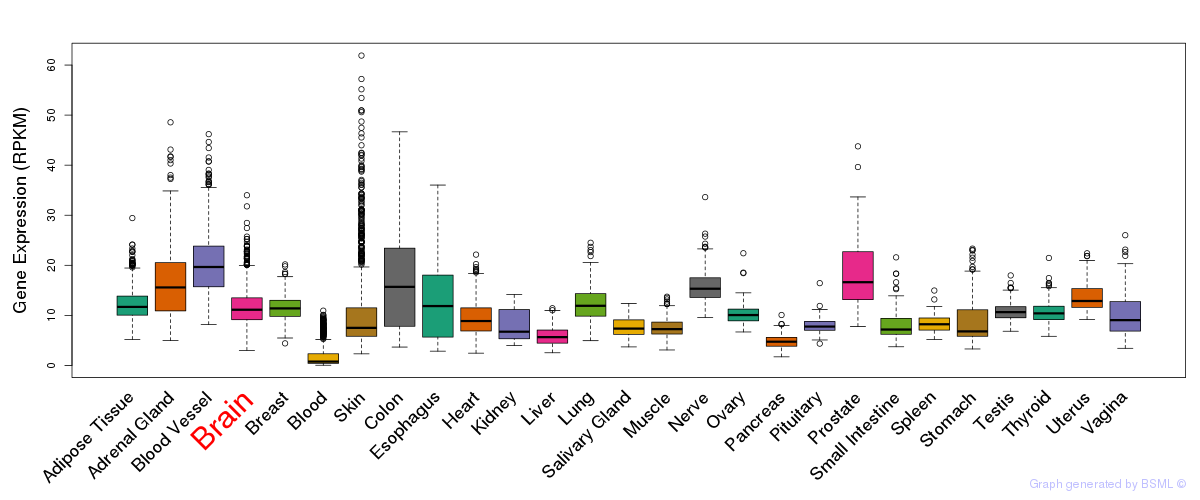
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C19orf25 | 0.88 | 0.83 |
| C19orf60 | 0.87 | 0.78 |
| MPG | 0.87 | 0.84 |
| C16orf13 | 0.86 | 0.76 |
| C19orf20 | 0.86 | 0.79 |
| SCAND1 | 0.85 | 0.79 |
| C9orf142 | 0.85 | 0.83 |
| GAMT | 0.84 | 0.81 |
| PHPT1 | 0.84 | 0.80 |
| DGCR6L | 0.84 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FLT1 | -0.37 | -0.40 |
| EEA1 | -0.37 | -0.32 |
| COBLL1 | -0.37 | -0.37 |
| CCDC55 | -0.37 | -0.35 |
| ZNHIT6 | -0.36 | -0.34 |
| CALD1 | -0.36 | -0.39 |
| ARHGAP5 | -0.36 | -0.33 |
| ITSN2 | -0.36 | -0.34 |
| AHNAK | -0.35 | -0.40 |
| UTRN | -0.35 | -0.34 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CEBPZ | CBF2 | HSP-CBF | NOC1 | CCAAT/enhancer binding protein (C/EBP), zeta | - | HPRD,BioGRID | 11306579 |
| CIITA | C2TA | CIITAIV | MHC2TA | NLRA | class II, major histocompatibility complex, transactivator | - | HPRD | 11003667 |
| CNTN2 | AXT | DKFZp781D102 | FLJ42746 | MGC157722 | TAG-1 | TAX | TAX1 | contactin 2 (axonal) | - | HPRD,BioGRID | 9032250 |
| CSDA | CSDA1 | DBPA | ZONAB | cold shock domain protein A | - | HPRD | 9442396 |
| DRAP1 | NC2-alpha | DR1-associated protein 1 (negative cofactor 2 alpha) | Two-hybrid | BioGRID | 16189514 |
| ELF1 | - | E74-like factor 1 (ets domain transcription factor) | - | HPRD | 9668064 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 11282029 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Myc interacts with NF-YB. This interaction was modeled on a demonstrated interaction between human Myc and mouse NF-YB | BIND | 11282029 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | - | HPRD,BioGRID | 8051128 |
| NFYC | CBF-C | CBFC | DKFZp667G242 | FLJ45775 | H1TF2A | HAP5 | HSM | NF-YC | nuclear transcription factor Y, gamma | - | HPRD | 9388234 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 9153318 |
| TP73 | P73 | tumor protein p73 | - | HPRD,BioGRID | 12167641 |
| YBX1 | BP-8 | CSDA2 | CSDB | DBPB | MDR-NF1 | MGC104858 | MGC110976 | MGC117250 | NSEP-1 | NSEP1 | YB-1 | YB1 | Y box binding protein 1 | - | HPRD | 7651426 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF GENES BY ATF4 | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS INDUCED BY AKT1 48HR DN | 27 | 17 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |