Gene Page: NHS
Summary ?
| GeneID | 4810 |
| Symbol | NHS |
| Synonyms | CTRCT40|CXN|SCML1 |
| Description | NHS actin remodeling regulator |
| Reference | MIM:300457|HGNC:HGNC:7820|Ensembl:ENSG00000188158|HPRD:02351|Vega:OTTHUMG00000022799 |
| Gene type | protein-coding |
| Map location | Xp22.13 |
| Sherlock p-value | 0.173 |
| TADA p-value | 0.021 |
| Fetal beta | -0.126 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NHS | chrX | 17744090 | G | A | NM_198270 | p.D601N | missense SNV | C | D | Schizophrenia | DNM:McCarthy_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16979498 | chr20 | 54771303 | NHS | 4810 | 0.18 | trans |
Section II. Transcriptome annotation
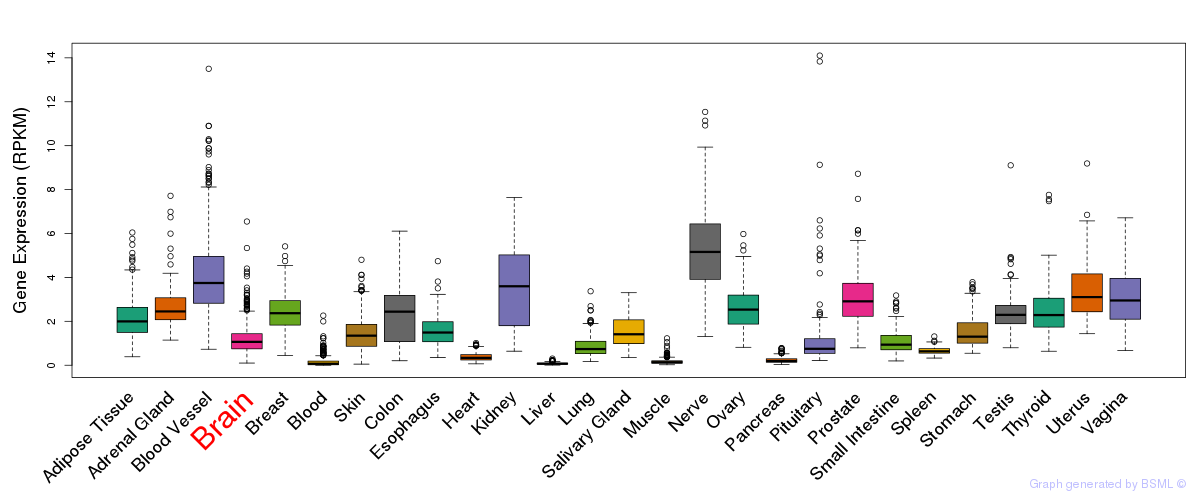
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KDM1 | 0.98 | 0.98 |
| FUBP1 | 0.98 | 0.98 |
| RBMX | 0.98 | 0.95 |
| XRN2 | 0.98 | 0.97 |
| KHDRBS1 | 0.97 | 0.98 |
| DHX9 | 0.97 | 0.97 |
| USP3 | 0.97 | 0.89 |
| ODC1 | 0.96 | 0.90 |
| EIF4B | 0.96 | 0.93 |
| MSH6 | 0.96 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.72 | -0.82 |
| C5orf53 | -0.71 | -0.78 |
| AIFM3 | -0.70 | -0.79 |
| AF347015.27 | -0.69 | -0.92 |
| AF347015.31 | -0.69 | -0.92 |
| S100B | -0.68 | -0.86 |
| PTH1R | -0.68 | -0.74 |
| AF347015.33 | -0.68 | -0.92 |
| MT-CO2 | -0.68 | -0.93 |
| FBXO2 | -0.68 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |