Gene Page: NPAS1
Summary ?
| GeneID | 4861 |
| Symbol | NPAS1 |
| Synonyms | MOP5|PASD5|bHLHe11 |
| Description | neuronal PAS domain protein 1 |
| Reference | MIM:603346|HGNC:HGNC:7894|Ensembl:ENSG00000130751|HPRD:09136|Vega:OTTHUMG00000183440 |
| Gene type | protein-coding |
| Map location | 19q13.32 |
| Pascal p-value | 0.23 |
| Sherlock p-value | 0.244 |
| Fetal beta | -0.312 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4698764 | chr4 | 111146783 | NPAS1 | 4861 | 0.04 | trans | ||
| rs17542473 | chr4 | 111182451 | NPAS1 | 4861 | 0.03 | trans | ||
| rs10054300 | chr5 | 51847657 | NPAS1 | 4861 | 0.07 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
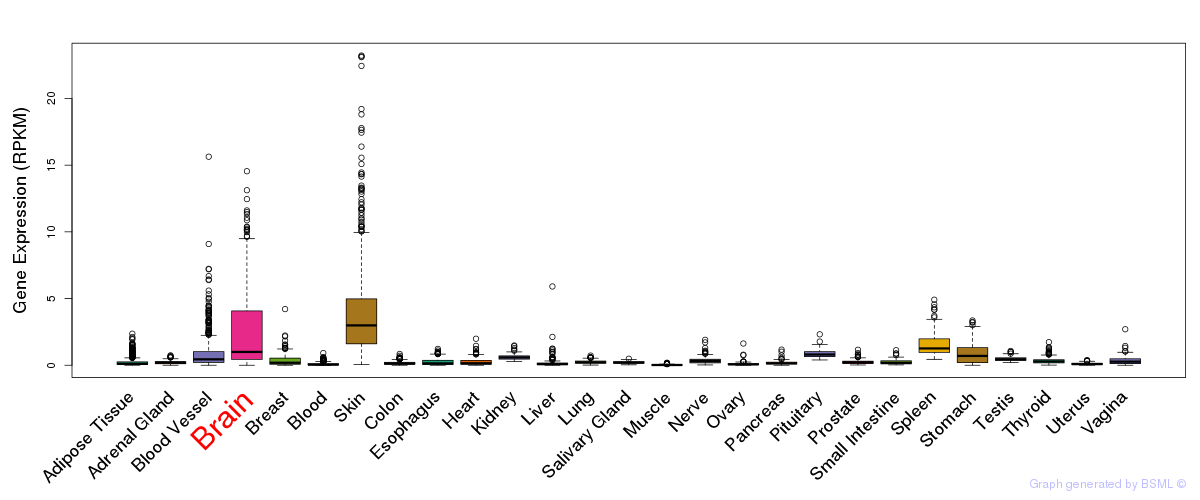
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NTNG1 | 0.72 | 0.48 |
| BTBD11 | 0.70 | 0.63 |
| FAM19A4 | 0.70 | 0.69 |
| TPH2 | 0.70 | 0.56 |
| CPNE9 | 0.69 | 0.69 |
| AC010087.3 | 0.69 | 0.64 |
| PTPN3 | 0.69 | 0.42 |
| SYT9 | 0.69 | 0.36 |
| DOK7 | 0.68 | 0.58 |
| NECAB2 | 0.68 | 0.67 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ISLR2 | -0.47 | -0.54 |
| PPAPDC1B | -0.47 | -0.56 |
| PLEKHO1 | -0.47 | -0.57 |
| ZNF300 | -0.46 | -0.50 |
| FOXG1B | -0.45 | -0.58 |
| CCNG2 | -0.45 | -0.53 |
| SF3A2 | -0.45 | -0.54 |
| ZNF551 | -0.45 | -0.54 |
| MYCN | -0.45 | -0.50 |
| AC004017.1 | -0.44 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 9012850 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 9012850 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007610 | behavior | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |