Gene Page: PAFAH1B3
Summary ?
| GeneID | 5050 |
| Symbol | PAFAH1B3 |
| Synonyms | PAFAHG |
| Description | platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| Reference | MIM:603074|HGNC:HGNC:8576|Ensembl:ENSG00000079462|HPRD:04354|Vega:OTTHUMG00000182795 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.095 |
| Sherlock p-value | 0.812 |
| Fetal beta | 2.468 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
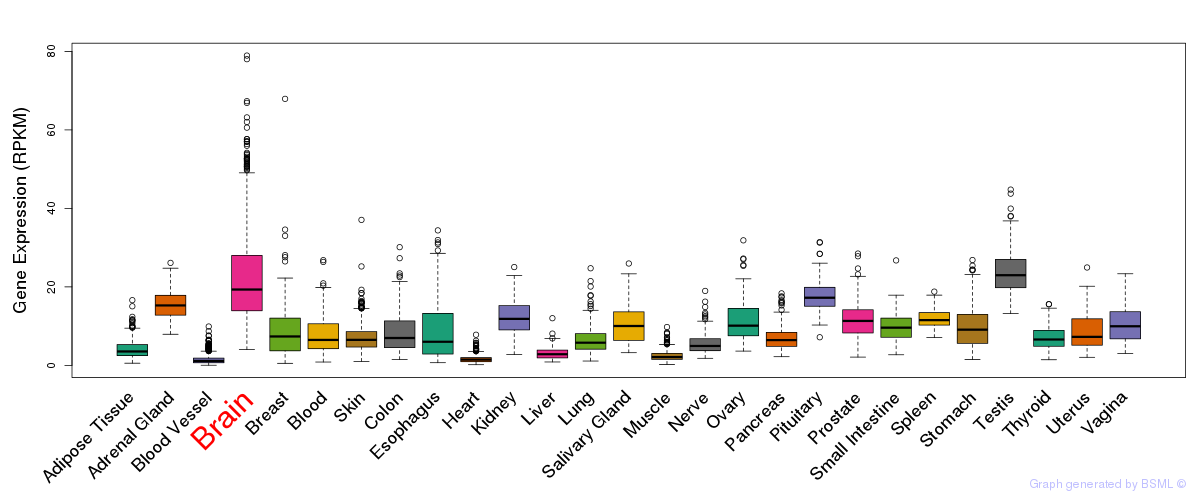
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMAD4 | 0.96 | 0.93 |
| STAG1 | 0.96 | 0.93 |
| SP3 | 0.96 | 0.94 |
| DHX9 | 0.96 | 0.93 |
| PPM1D | 0.95 | 0.94 |
| ZFP161 | 0.95 | 0.94 |
| HNRNPK | 0.95 | 0.93 |
| SFRS1 | 0.95 | 0.94 |
| ALDH18A1 | 0.95 | 0.93 |
| CORO1C | 0.95 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.70 | -0.87 |
| MT-CO2 | -0.70 | -0.86 |
| AF347015.31 | -0.69 | -0.85 |
| IFI27 | -0.69 | -0.86 |
| HLA-F | -0.69 | -0.76 |
| AF347015.33 | -0.68 | -0.82 |
| AF347015.27 | -0.67 | -0.82 |
| C5orf53 | -0.67 | -0.71 |
| PTH1R | -0.67 | -0.75 |
| MT-CYB | -0.66 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 16189514 | |
| GO:0016788 | hydrolase activity, acting on ester bonds | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 7669037 |
| GO:0016042 | lipid catabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRD7 | BP75 | CELTIX1 | NAG4 | bromodomain containing 7 | Two-hybrid | BioGRID | 16169070 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| C7orf64 | DKFZP564O0523 | DKFZp686D1651 | HSPC304 | chromosome 7 open reading frame 64 | Two-hybrid | BioGRID | 16169070 |
| CCDC90B | MDS011 | MDS025 | MGC104239 | coiled-coil domain containing 90B | Two-hybrid | BioGRID | 16169070 |
| CHMP7 | MGC29816 | CHMP family, member 7 | Two-hybrid | BioGRID | 16189514 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| ECH1 | HPXEL | enoyl Coenzyme A hydratase 1, peroxisomal | Two-hybrid | BioGRID | 16169070 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | Two-hybrid | BioGRID | 16169070 |
| GBP2 | - | guanylate binding protein 2, interferon-inducible | Two-hybrid | BioGRID | 16169070 |
| GDF9 | - | growth differentiation factor 9 | Two-hybrid | BioGRID | 16169070 |
| HSPH1 | DKFZp686M05240 | HSP105 | HSP105A | HSP105B | KIAA0201 | NY-CO-25 | heat shock 105kDa/110kDa protein 1 | Reconstituted Complex | BioGRID | 14733918 |
| IGSF21 | FLJ41177 | MGC15730 | immunoglobin superfamily, member 21 | Two-hybrid | BioGRID | 16169070 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B1 | LIS1 | LIS2 | MDCR | MDS | PAFAH | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | - | HPRD,BioGRID | 10727864 |
| PAFAH1B2 | - | platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa | Two-hybrid | BioGRID | 16189514 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16189514 |
| PLEKHM1 | AP162 | B2 | KIAA0356 | OPTB6 | pleckstrin homology domain containing, family M (with RUN domain) member 1 | Two-hybrid | BioGRID | 16169070 |
| SDCBP2 | FLJ12256 | SITAC18 | ST-2 | syndecan binding protein (syntenin) 2 | Two-hybrid | BioGRID | 16189514 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Two-hybrid | BioGRID | 16169070 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | Two-hybrid | BioGRID | 16169070 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | Two-hybrid | BioGRID | 16169070 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16169070 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ETHER LIPID METABOLISM | 33 | 15 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| CREIGHTON AKT1 SIGNALING VIA MTOR DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |