| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR UP
| 85 | 54 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP
| 182 | 110 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP
| 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP
| 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP
| 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP
| 418 | 263 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP
| 341 | 197 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY DN
| 29 | 21 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP
| 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN
| 141 | 84 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP
| 536 | 340 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HEMGN
| 31 | 21 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR
| 43 | 35 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN
| 514 | 319 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN
| 203 | 134 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN
| 584 | 395 | All SZGR 2.0 genes in this pathway |
| COWLING MYCN TARGETS
| 43 | 27 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP
| 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC DN
| 59 | 35 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS
| 78 | 45 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP
| 79 | 58 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP
| 140 | 83 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN
| 163 | 115 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP
| 183 | 111 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN
| 48 | 26 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP
| 70 | 47 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN
| 86 | 67 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70
| 67 | 33 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS
| 78 | 52 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN
| 49 | 29 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN
| 57 | 39 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING SCAR UP
| 30 | 21 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP
| 198 | 132 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7
| 51 | 34 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP
| 268 | 157 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5
| 46 | 36 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN
| 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN
| 540 | 340 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS
| 78 | 49 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP
| 43 | 34 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH FLT3 ITD
| 40 | 22 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION
| 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED
| 351 | 238 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN
| 56 | 39 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP
| 194 | 133 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP
| 271 | 165 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN
| 448 | 282 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
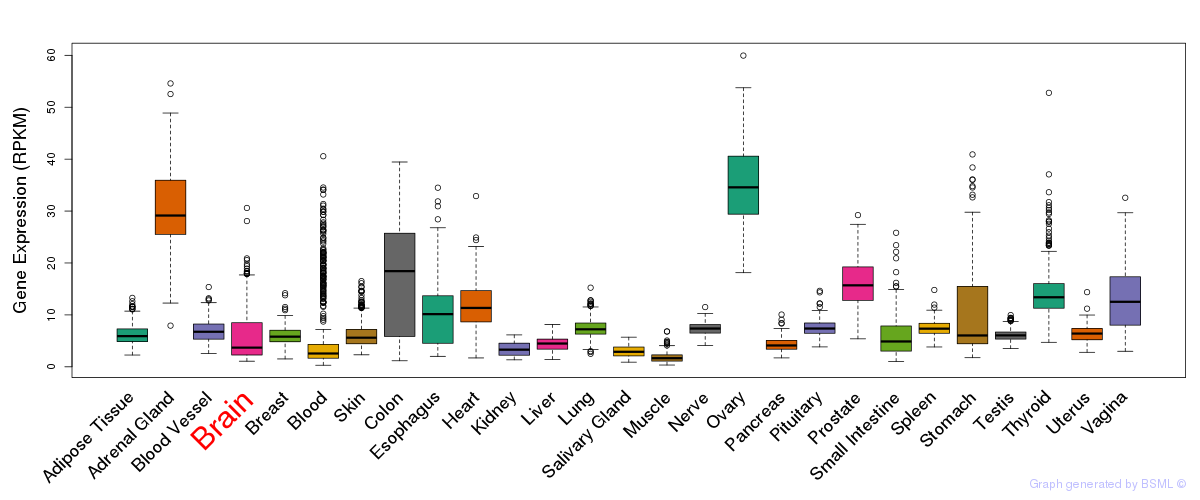
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation