Gene Page: PCBD1
Summary ?
| GeneID | 5092 |
| Symbol | PCBD1 |
| Synonyms | DCOH|PCBD|PCD|PHS |
| Description | pterin-4 alpha-carbinolamine dehydratase 1 |
| Reference | MIM:126090|HGNC:HGNC:8646|Ensembl:ENSG00000166228|HPRD:00523|Vega:OTTHUMG00000018417 |
| Gene type | protein-coding |
| Map location | 10q22 |
| Pascal p-value | 0.793 |
| Sherlock p-value | 0.015 |
| Fetal beta | -0.635 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
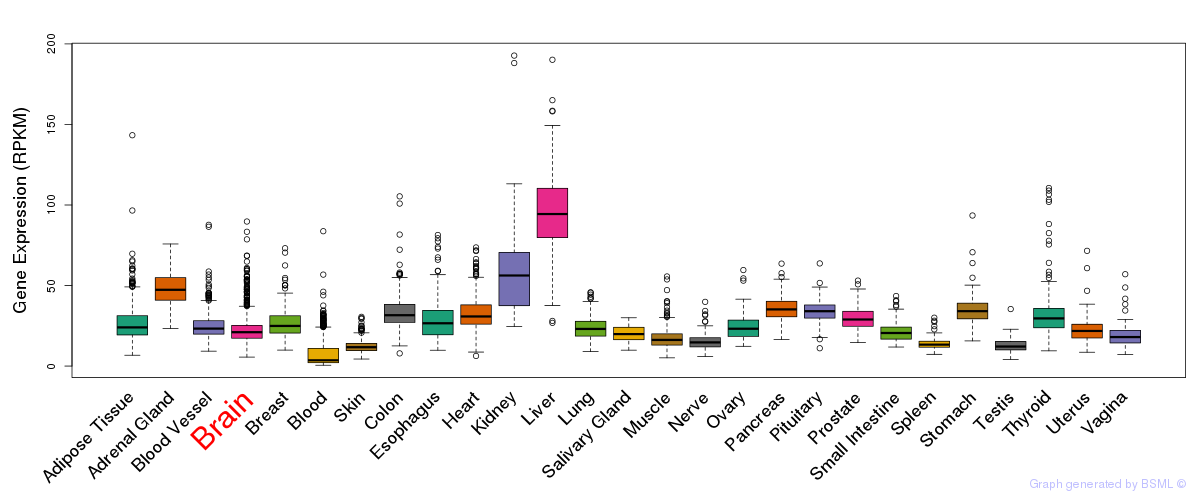
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CACNA1C | CACH2 | CACN2 | CACNL1A1 | CCHL1A1 | CaV1.2 | MGC120730 | TS | calcium channel, voltage-dependent, L type, alpha 1C subunit | - | HPRD,BioGRID | 11461190 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| DYRK1B | MIRK | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | - | HPRD,BioGRID | 11980910 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| GORASP2 | DKFZp434D156 | FLJ13139 | GOLPH6 | GRASP55 | GRS2 | p59 | golgi reassembly stacking protein 2, 55kDa | Two-hybrid | BioGRID | 16189514 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 9092652 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| NIF3L1 | ALS2CR1 | CALS-7 | MDS015 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | Two-hybrid | BioGRID | 16189514 |
| NUDT18 | FLJ22494 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | Two-hybrid | BioGRID | 16189514 |
| PAH | PH | PKU | PKU1 | phenylalanine hydroxylase | Two-hybrid | BioGRID | 11461190 |
| PCBD1 | DCOH | PCBD | PCD | PHS | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| WDYHV1 | C8orf32 | FLJ10204 | WDYHV motif containing 1 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 MCF10A | 43 | 28 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 60 MCF10A | 57 | 42 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |