Gene Page: MED31
Summary ?
| GeneID | 51003 |
| Symbol | MED31 |
| Synonyms | 3110004H13Rik|CGI-125|Soh1 |
| Description | mediator complex subunit 31 |
| Reference | HGNC:HGNC:24260|Ensembl:ENSG00000108590|HPRD:14382|Vega:OTTHUMG00000102051 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.13 |
| Fetal beta | -0.162 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26623286 | 17 | 6555130 | MED31 | 1.84E-8 | -0.012 | 6.57E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
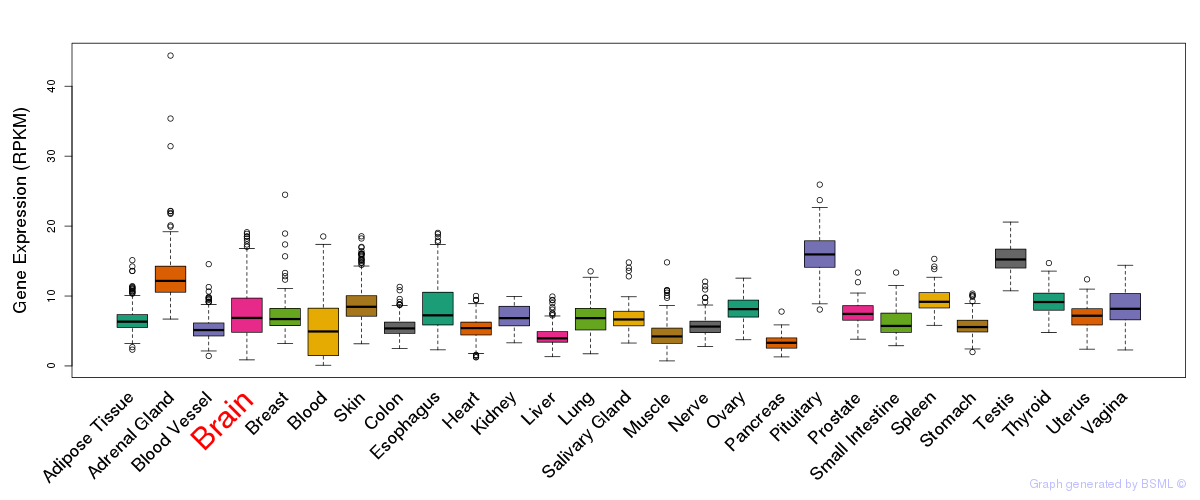
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Two-hybrid | BioGRID | 16169070 |
| AGR2 | AG2 | GOB-4 | HAG-2 | XAG-2 | anterior gradient homolog 2 (Xenopus laevis) | Two-hybrid | BioGRID | 16169070 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| C2orf44 | FLJ21945 | PP384 | chromosome 2 open reading frame 44 | Two-hybrid | BioGRID | 16169070 |
| C8orf30A | FLJ40907 | chromosome 8 open reading frame 30A | Two-hybrid | BioGRID | 16169070 |
| CACNB4 | CAB4 | CACNLB4 | EA5 | EJM | calcium channel, voltage-dependent, beta 4 subunit | Two-hybrid | BioGRID | 16169070 |
| CCT7 | CCT-ETA | Ccth | MGC110985 | Nip7-1 | TCP-1-eta | chaperonin containing TCP1, subunit 7 (eta) | Two-hybrid | BioGRID | 16169070 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-MS | BioGRID | 10198638 |
| CPE | - | carboxypeptidase E | Two-hybrid | BioGRID | 16169070 |
| DLEU1 | BCMS | DLB1 | DLEU2 | LEU1 | LEU2 | MGC22430 | NCRNA00021 | XTP6 | deleted in lymphocytic leukemia 1 (non-protein coding) | Two-hybrid | BioGRID | 16169070 |
| DNM1 | DNM | dynamin 1 | Two-hybrid | BioGRID | 16169070 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| EGR2 | AT591 | CMT1D | CMT4E | DKFZp686J1957 | FLJ14547 | KROX20 | early growth response 2 (Krox-20 homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| EPN1 | - | epsin 1 | Two-hybrid | BioGRID | 16169070 |
| ERH | DROER | FLJ27340 | enhancer of rudimentary homolog (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD | 15383276 |
| GSTM4 | GSTM4-4 | GTM4 | MGC131945 | MGC9247 | glutathione S-transferase mu 4 | Two-hybrid | BioGRID | 16169070 |
| HMOX2 | HO-2 | heme oxygenase (decycling) 2 | Two-hybrid | BioGRID | 16169070 |
| HNRNPUL1 | E1B-AP5 | E1BAP5 | FLJ12944 | HNRPUL1 | heterogeneous nuclear ribonucleoprotein U-like 1 | Two-hybrid | BioGRID | 16169070 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | Two-hybrid | BioGRID | 16169070 |
| HSPB3 | HSPL27 | heat shock 27kDa protein 3 | Two-hybrid | BioGRID | 16169070 |
| HTT | HD | IT15 | huntingtin | - | HPRD | 15383276 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| LYPLA2 | APT-2 | DJ886K2.4 | lysophospholipase II | Two-hybrid | BioGRID | 16169070 |
| MAFG | MGC13090 | MGC20149 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) | Two-hybrid | BioGRID | 16169070 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14576168 |15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | - | HPRD,BioGRID | 12584197 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD,BioGRID | 14576168 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Two-hybrid | BioGRID | 16169070 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | Two-hybrid | BioGRID | 16169070 |
| ORAI2 | C7orf19 | CBCIP2 | FLJ12474 | FLJ14733 | FLJ44818 | TMEM142B | ORAI calcium release-activated calcium modulator 2 | Two-hybrid | BioGRID | 16169070 |
| PABPC4 | APP-1 | APP1 | FLJ43938 | PABP4 | iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) | - | HPRD | 15383276 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PFDN1 | PDF | PFD1 | prefoldin subunit 1 | Two-hybrid | BioGRID | 16169070 |
| PMF1 | - | polyamine-modulated factor 1 | Two-hybrid | BioGRID | 16169070 |
| PQBP1 | MRX55 | MRXS3 | MRXS8 | NPW38 | RENS1 | SHS | polyglutamine binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| RAB25 | CATX-8 | RAB25, member RAS oncogene family | - | HPRD | 11591653 |
| RBM23 | CAPERbeta | FLJ10482 | MGC4458 | PP239 | RNPC4 | RNA binding motif protein 23 | Two-hybrid | BioGRID | 16169070 |
| RFC5 | MGC1155 | RFC36 | replication factor C (activator 1) 5, 36.5kDa | Two-hybrid | BioGRID | 16169070 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SELENBP1 | FLJ13813 | LPSB | SP56 | hSBP | hSP56 | selenium binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| TGIF1 | HPE4 | MGC39747 | MGC5066 | TGIF | TGFB-induced factor homeobox 1 | Two-hybrid | BioGRID | 16169070 |
| TINAGL1 | ARG1 | LCN7 | LIECG3 | TINAGRP | tubulointerstitial nephritis antigen-like 1 | Two-hybrid | BioGRID | 16169070 |
| UBE2B | E2-17kDa | HHR6B | HR6B | RAD6B | UBC2 | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | Two-hybrid | BioGRID | 16169070 |
| ZBTB45 | DKFZp547H249 | FLJ14486 | ZNF499 | zinc finger and BTB domain containing 45 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| CARDOSO RESPONSE TO GAMMA RADIATION AND 3AB | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 DN | 63 | 39 | All SZGR 2.0 genes in this pathway |