| KEGG GLYCOLYSIS GLUCONEOGENESIS
| 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG CITRATE CYCLE TCA CYCLE
| 32 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG PYRUVATE METABOLISM
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG PPAR SIGNALING PATHWAY
| 69 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY
| 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY
| 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG PROXIMAL TUBULE BICARBONATE RECLAMATION
| 23 | 16 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY
| 45 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY
| 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME ABACAVIR TRANSPORT AND METABOLISM
| 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCONEOGENESIS
| 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION
| 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM
| 69 | 42 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP
| 108 | 67 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN
| 291 | 176 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE
| 217 | 143 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND
| 69 | 40 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON
| 149 | 76 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN
| 108 | 84 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN
| 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN
| 65 | 44 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN
| 200 | 112 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP
| 398 | 262 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP
| 205 | 126 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN
| 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN
| 74 | 45 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN
| 63 | 40 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN
| 546 | 351 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN
| 64 | 42 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP
| 555 | 346 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES
| 244 | 154 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN
| 48 | 34 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE UP
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP
| 314 | 201 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN
| 86 | 66 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL
| 254 | 164 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN
| 84 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN
| 504 | 323 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP
| 17 | 11 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN
| 434 | 302 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3
| 196 | 93 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS
| 245 | 148 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP
| 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP
| 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP
| 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN
| 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS
| 425 | 261 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP
| 185 | 112 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES
| 31 | 26 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP
| 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN
| 435 | 289 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN
| 179 | 97 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN
| 113 | 76 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3
| 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN
| 267 | 160 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN
| 80 | 51 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 5
| 30 | 22 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS
| 32 | 23 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP
| 857 | 456 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 5 DN
| 50 | 31 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS UP
| 21 | 16 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2
| 98 | 67 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS UP
| 51 | 39 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
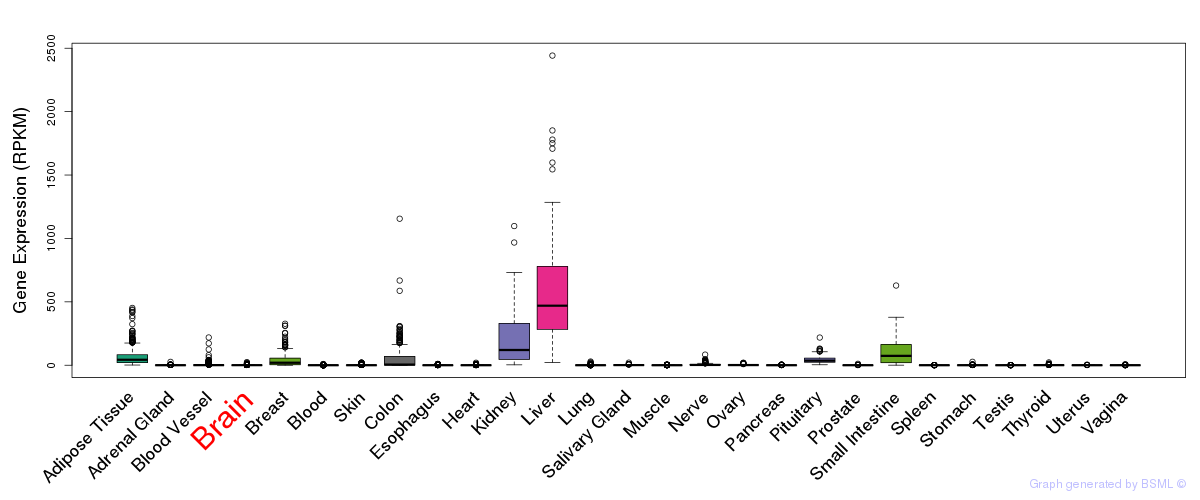
 eQTL annotation
eQTL annotation