Gene Page: COPS4
Summary ?
| GeneID | 51138 |
| Symbol | COPS4 |
| Synonyms | CSN4 |
| Description | COP9 signalosome subunit 4 |
| Reference | MIM:616008|HGNC:HGNC:16702|Ensembl:ENSG00000138663|HPRD:09888|Vega:OTTHUMG00000130298 |
| Gene type | protein-coding |
| Map location | 4q21.22 |
| Pascal p-value | 0.033 |
| Sherlock p-value | 0.475 |
| Fetal beta | -0.255 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24137332 | 4 | 83957095 | COPS4 | 2.962E-4 | 0.535 | 0.039 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
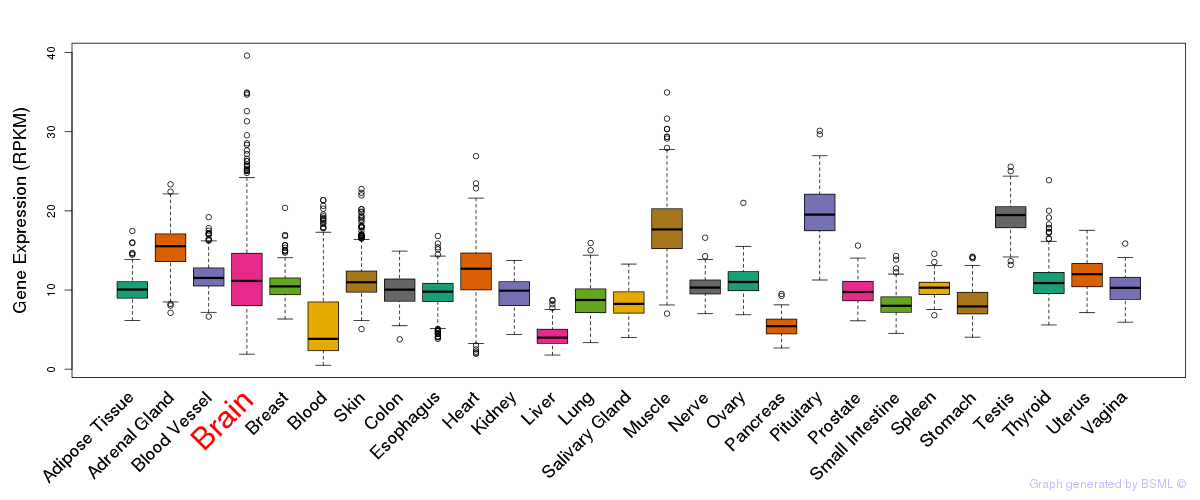
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C19orf57 | MGC11271 | MGC149720 | chromosome 19 open reading frame 57 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | CSN2 interacts with CSN4. | BIND | 12615944 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | CSN4 interacts with CSN3. | BIND | 12615944 |
| COPS3 | SGN3 | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS4 | MGC10899 | MGC15160 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | CSN4 interacts with itself. | BIND | 12615944 |
| COPS5 | CSN5 | JAB1 | MGC3149 | MOV-34 | SGN5 | COP9 constitutive photomorphogenic homolog subunit 5 (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | CSN4 interacts with CSN6. | BIND | 12615944 |
| COPS7A | MGC110877 | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | - | HPRD | 9707402 |14647295 |
| COPS7A | MGC110877 | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | CSN4 interacts with CSN7. | BIND | 12615944 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | CSN4 interacts with CSN8. | BIND | 12615944 |
| COPS8 | COP9 | CSN8 | MGC1297 | MGC43256 | SGN8 | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | - | HPRD,BioGRID | 9707402 |14647295 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | - | HPRD | 9707402 |14647295 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | CSN1 interacts with CSN4. | BIND | 12615944 |
| GPS1 | COPS1 | CSN1 | MGC71287 | G protein pathway suppressor 1 | - | HPRD | 11114242 |
| MBIP | - | MAP3K12 binding inhibitory protein 1 | Two-hybrid | BioGRID | 16189514 |
| RCBTB2 | CHC1L | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 | Two-hybrid | BioGRID | 16189514 |
| USHBP1 | AIEBP | FLJ38709 | FLJ90681 | MCC2 | Usher syndrome 1C binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| WDR8 | FLJ20430 | MGC99569 | WD repeat domain 8 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE TUMOR SAMPLE UP | 16 | 12 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |