Gene Page: ACP6
Summary ?
| GeneID | 51205 |
| Symbol | ACP6 |
| Synonyms | ACPL1|LPAP|PACPL1 |
| Description | acid phosphatase 6, lysophosphatidic |
| Reference | MIM:611471|HGNC:HGNC:29609|HPRD:16472| |
| Gene type | protein-coding |
| Map location | 1q21 |
| Sherlock p-value | 0.892 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27244432 | 1 | 147142282 | ACP6 | 8.52E-9 | -0.008 | 3.98E-6 | DMG:Jaffe_2016 |
| cg05968052 | 1 | 147142184 | ACP6 | 3.28E-8 | -0.01 | 9.87E-6 | DMG:Jaffe_2016 |
| cg18709479 | 1 | 147142473 | ACP6 | 9.41E-8 | -0.006 | 2.13E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3820129 | chr1 | 147091971 | ACP6 | 51205 | 0.04 | cis | ||
| rs12037951 | chr1 | 147102853 | ACP6 | 51205 | 0.13 | cis | ||
| rs1344 | chr1 | 147119272 | ACP6 | 51205 | 4.311E-4 | cis | ||
| rs6674938 | chr1 | 147131464 | ACP6 | 51205 | 0.01 | cis | ||
| rs11579007 | chr1 | 147136283 | ACP6 | 51205 | 0 | cis | ||
| rs12123144 | chr1 | 76550208 | ACP6 | 51205 | 0.1 | trans | ||
| rs1344 | chr1 | 147119272 | ACP6 | 51205 | 0.04 | trans | ||
| rs11579007 | chr1 | 147136283 | ACP6 | 51205 | 0.08 | trans | ||
| rs1467414 | chr20 | 50243844 | ACP6 | 51205 | 0.13 | trans | ||
| rs597577 | 1 | 147088907 | ACP6 | ENSG00000162836.7 | 2.682E-6 | 0 | 53711 | gtex_brain_ba24 |
| rs2236570 | 1 | 147093887 | ACP6 | ENSG00000162836.7 | 3.555E-7 | 0 | 48731 | gtex_brain_ba24 |
| rs17361839 | 1 | 147099688 | ACP6 | ENSG00000162836.7 | 2.789E-6 | 0 | 42930 | gtex_brain_ba24 |
| rs12037951 | 1 | 147102854 | ACP6 | ENSG00000162836.7 | 2.694E-6 | 0 | 39764 | gtex_brain_ba24 |
| rs10494247 | 1 | 147113387 | ACP6 | ENSG00000162836.7 | 6.602E-8 | 0 | 29231 | gtex_brain_ba24 |
| rs11811290 | 1 | 147114285 | ACP6 | ENSG00000162836.7 | 1.005E-6 | 0 | 28333 | gtex_brain_ba24 |
| rs10900387 | 1 | 147114303 | ACP6 | ENSG00000162836.7 | 8.046E-8 | 0 | 28315 | gtex_brain_ba24 |
| rs12045115 | 1 | 147118181 | ACP6 | ENSG00000162836.7 | 2.128E-11 | 0 | 24437 | gtex_brain_ba24 |
| rs12744461 | 1 | 147118500 | ACP6 | ENSG00000162836.7 | 2.67E-6 | 0 | 24118 | gtex_brain_ba24 |
| rs1344 | 1 | 147119273 | ACP6 | ENSG00000162836.7 | 4.847E-10 | 0 | 23345 | gtex_brain_ba24 |
| rs11577090 | 1 | 147122616 | ACP6 | ENSG00000162836.7 | 4.94E-6 | 0 | 20002 | gtex_brain_ba24 |
| rs946904 | 1 | 147122831 | ACP6 | ENSG00000162836.7 | 4.94E-6 | 0 | 19787 | gtex_brain_ba24 |
| rs2153463 | 1 | 147124310 | ACP6 | ENSG00000162836.7 | 1.636E-10 | 0 | 18308 | gtex_brain_ba24 |
| rs9660040 | 1 | 147124726 | ACP6 | ENSG00000162836.7 | 2.551E-9 | 0 | 17892 | gtex_brain_ba24 |
| rs28700004 | 1 | 147124960 | ACP6 | ENSG00000162836.7 | 5.738E-11 | 0 | 17658 | gtex_brain_ba24 |
| rs4950465 | 1 | 147125235 | ACP6 | ENSG00000162836.7 | 3.658E-8 | 0 | 17383 | gtex_brain_ba24 |
| rs12119079 | 1 | 147129014 | ACP6 | ENSG00000162836.7 | 5.24E-11 | 0 | 13604 | gtex_brain_ba24 |
| rs12145385 | 1 | 147129786 | ACP6 | ENSG00000162836.7 | 5.24E-11 | 0 | 12832 | gtex_brain_ba24 |
| rs3766511 | 1 | 147130043 | ACP6 | ENSG00000162836.7 | 1.566E-6 | 0 | 12575 | gtex_brain_ba24 |
| rs3766508 | 1 | 147130355 | ACP6 | ENSG00000162836.7 | 4.296E-7 | 0 | 12263 | gtex_brain_ba24 |
| rs33965274 | 1 | 147130661 | ACP6 | ENSG00000162836.7 | 1.499E-6 | 0 | 11957 | gtex_brain_ba24 |
| rs6674938 | 1 | 147131465 | ACP6 | ENSG00000162836.7 | 2.643E-9 | 0 | 11153 | gtex_brain_ba24 |
| rs34675044 | 1 | 147131501 | ACP6 | ENSG00000162836.7 | 2.774E-6 | 0 | 11117 | gtex_brain_ba24 |
| rs2275552 | 1 | 147131945 | ACP6 | ENSG00000162836.7 | 2.639E-9 | 0 | 10673 | gtex_brain_ba24 |
| rs140291349 | 1 | 147135406 | ACP6 | ENSG00000162836.7 | 4.771E-9 | 0 | 7212 | gtex_brain_ba24 |
| rs3820129 | 1 | 147091972 | ACP6 | ENSG00000162836.7 | 4.866E-8 | 0 | 50646 | gtex_brain_putamen_basal |
| rs72506255 | 1 | 147095064 | ACP6 | ENSG00000162836.7 | 1.991E-6 | 0 | 47554 | gtex_brain_putamen_basal |
| rs12082988 | 1 | 147099315 | ACP6 | ENSG00000162836.7 | 4.232E-8 | 0 | 43303 | gtex_brain_putamen_basal |
| rs12085690 | 1 | 147100887 | ACP6 | ENSG00000162836.7 | 4.487E-8 | 0 | 41731 | gtex_brain_putamen_basal |
| rs12037951 | 1 | 147102854 | ACP6 | ENSG00000162836.7 | 1.939E-7 | 0 | 39764 | gtex_brain_putamen_basal |
| rs12085449 | 1 | 147103354 | ACP6 | ENSG00000162836.7 | 3.591E-8 | 0 | 39264 | gtex_brain_putamen_basal |
| rs11587616 | 1 | 147109122 | ACP6 | ENSG00000162836.7 | 3.613E-8 | 0 | 33496 | gtex_brain_putamen_basal |
| rs10494247 | 1 | 147113387 | ACP6 | ENSG00000162836.7 | 1.49E-8 | 0 | 29231 | gtex_brain_putamen_basal |
| rs11811290 | 1 | 147114285 | ACP6 | ENSG00000162836.7 | 2.265E-7 | 0 | 28333 | gtex_brain_putamen_basal |
| rs10900387 | 1 | 147114303 | ACP6 | ENSG00000162836.7 | 1.437E-8 | 0 | 28315 | gtex_brain_putamen_basal |
| rs12045115 | 1 | 147118181 | ACP6 | ENSG00000162836.7 | 2.024E-11 | 0 | 24437 | gtex_brain_putamen_basal |
| rs12744461 | 1 | 147118500 | ACP6 | ENSG00000162836.7 | 4.546E-11 | 0 | 24118 | gtex_brain_putamen_basal |
| rs1344 | 1 | 147119273 | ACP6 | ENSG00000162836.7 | 1.442E-11 | 0 | 23345 | gtex_brain_putamen_basal |
| rs11577090 | 1 | 147122616 | ACP6 | ENSG00000162836.7 | 6.899E-10 | 0 | 20002 | gtex_brain_putamen_basal |
| rs946904 | 1 | 147122831 | ACP6 | ENSG00000162836.7 | 6.899E-10 | 0 | 19787 | gtex_brain_putamen_basal |
| rs2153463 | 1 | 147124310 | ACP6 | ENSG00000162836.7 | 9.55E-12 | 0 | 18308 | gtex_brain_putamen_basal |
| rs9660040 | 1 | 147124726 | ACP6 | ENSG00000162836.7 | 2.483E-11 | 0 | 17892 | gtex_brain_putamen_basal |
| rs28700004 | 1 | 147124960 | ACP6 | ENSG00000162836.7 | 2.911E-11 | 0 | 17658 | gtex_brain_putamen_basal |
| rs4950465 | 1 | 147125235 | ACP6 | ENSG00000162836.7 | 2.208E-9 | 0 | 17383 | gtex_brain_putamen_basal |
| rs4950466 | 1 | 147125498 | ACP6 | ENSG00000162836.7 | 2.277E-9 | 0 | 17120 | gtex_brain_putamen_basal |
| rs12119079 | 1 | 147129014 | ACP6 | ENSG00000162836.7 | 2.818E-11 | 0 | 13604 | gtex_brain_putamen_basal |
| rs12145385 | 1 | 147129786 | ACP6 | ENSG00000162836.7 | 2.818E-11 | 0 | 12832 | gtex_brain_putamen_basal |
| rs7518236 | 1 | 147130000 | ACP6 | ENSG00000162836.7 | 9.997E-8 | 0 | 12618 | gtex_brain_putamen_basal |
| rs3766511 | 1 | 147130043 | ACP6 | ENSG00000162836.7 | 4.336E-11 | 0 | 12575 | gtex_brain_putamen_basal |
| rs3766509 | 1 | 147130275 | ACP6 | ENSG00000162836.7 | 6.333E-9 | 0 | 12343 | gtex_brain_putamen_basal |
| rs3766508 | 1 | 147130355 | ACP6 | ENSG00000162836.7 | 2.492E-10 | 0 | 12263 | gtex_brain_putamen_basal |
| rs33965274 | 1 | 147130661 | ACP6 | ENSG00000162836.7 | 4.603E-11 | 0 | 11957 | gtex_brain_putamen_basal |
| rs6674938 | 1 | 147131465 | ACP6 | ENSG00000162836.7 | 1.085E-11 | 0 | 11153 | gtex_brain_putamen_basal |
| rs34675044 | 1 | 147131501 | ACP6 | ENSG00000162836.7 | 6.39E-11 | 0 | 11117 | gtex_brain_putamen_basal |
| rs2275552 | 1 | 147131945 | ACP6 | ENSG00000162836.7 | 9.55E-12 | 0 | 10673 | gtex_brain_putamen_basal |
| rs140291349 | 1 | 147135406 | ACP6 | ENSG00000162836.7 | 5.956E-11 | 0 | 7212 | gtex_brain_putamen_basal |
| rs11579007 | 1 | 147136284 | ACP6 | ENSG00000162836.7 | 1.836E-9 | 0 | 6334 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
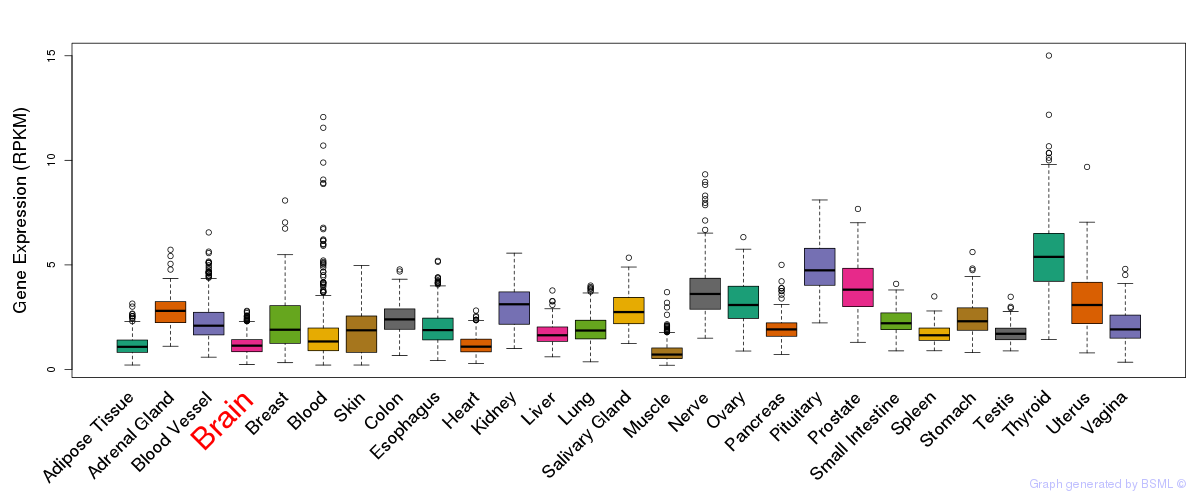
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003993 | acid phosphatase activity | TAS | 10506173 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006629 | lipid metabolic process | TAS | 10506173 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 10506173 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIBOFLAVIN METABOLISM | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |