Gene Page: CSAD
Summary ?
| GeneID | 51380 |
| Symbol | CSAD |
| Synonyms | CSD|PCAP |
| Description | cysteine sulfinic acid decarboxylase |
| Reference | MIM:616569|HGNC:HGNC:18966|Ensembl:ENSG00000139631|HPRD:16763|Vega:OTTHUMG00000048076 |
| Gene type | protein-coding |
| Map location | 12q13.11-q14.3 |
| Pascal p-value | 0.268 |
| Sherlock p-value | 0.446 |
| Fetal beta | 0.545 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02710461 | 12 | 53566978 | CSAD | 7.01E-5 | 0.315 | 0.024 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11170324 | chr12 | 53290402 | CSAD | 51380 | 0.04 | cis | ||
| rs2716734 | chr2 | 39947720 | CSAD | 51380 | 0 | trans | ||
| rs2716736 | chr2 | 39947946 | CSAD | 51380 | 0 | trans | ||
| rs2597796 | chr4 | 17535859 | CSAD | 51380 | 0.09 | trans | ||
| rs1587434 | chr6 | 66615354 | CSAD | 51380 | 0.03 | trans | ||
| rs17165823 | chr7 | 93476735 | CSAD | 51380 | 0.14 | trans | ||
| rs2294031 | chr8 | 1812077 | CSAD | 51380 | 0.11 | trans | ||
| rs12255258 | chr10 | 106306927 | CSAD | 51380 | 0.01 | trans | ||
| rs1241940 | chr14 | 84008851 | CSAD | 51380 | 0.03 | trans | ||
| rs12895787 | chr14 | 84017913 | CSAD | 51380 | 0.13 | trans | ||
| rs349973 | chr17 | 50642288 | CSAD | 51380 | 0.13 | trans | ||
| rs11170398 | 12 | 53471839 | CSAD | ENSG00000139631.14 | 2.418E-6 | 0.04 | 103296 | gtex_brain_putamen_basal |
| rs11170400 | 12 | 53472002 | CSAD | ENSG00000139631.14 | 2.363E-6 | 0.04 | 103133 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
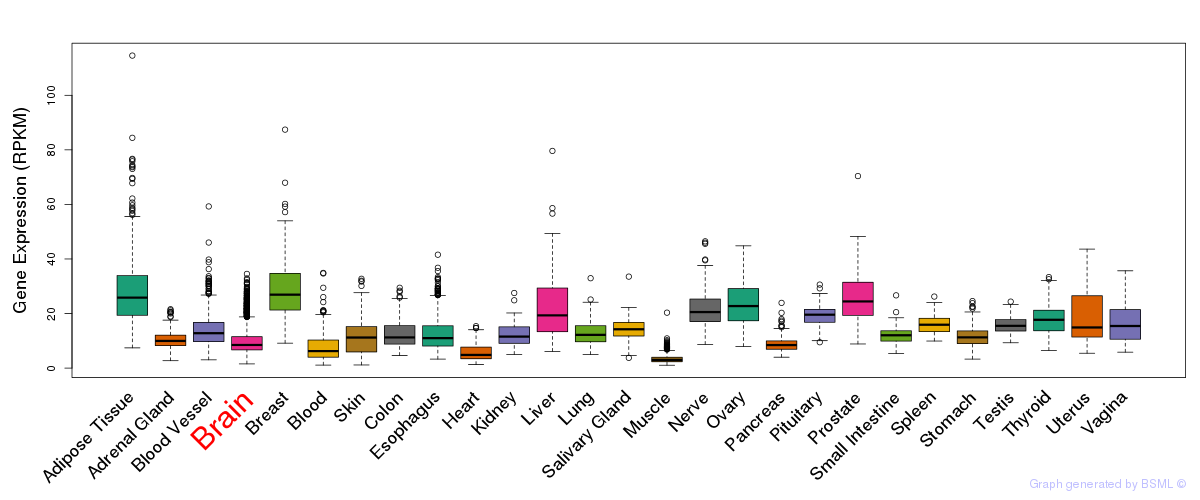
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TAURINE AND HYPOTAURINE METABOLISM | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME SULFUR AMINO ACID METABOLISM | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| PATTERSON DOCETAXEL RESISTANCE | 29 | 20 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| CARDOSO RESPONSE TO GAMMA RADIATION AND 3AB | 18 | 11 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |