Gene Page: PDE4A
Summary ?
| GeneID | 5141 |
| Symbol | PDE4A |
| Synonyms | DPDE2|PDE4|PDE46 |
| Description | phosphodiesterase 4A |
| Reference | MIM:600126|HGNC:HGNC:8780|Ensembl:ENSG00000065989|HPRD:02527|Vega:OTTHUMG00000180411 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.612 |
| Sherlock p-value | 0.978 |
| Fetal beta | -1.803 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19355182 | 19 | 10543010 | PDE4A | 8.41E-5 | -0.571 | 0.026 | DMG:Wockner_2014 |
| cg14536784 | 19 | 10542623 | PDE4A | 4.132E-4 | -0.32 | 0.044 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6924957 | chr6 | 96853578 | PDE4A | 5141 | 0.15 | trans | ||
| rs6568383 | 0 | PDE4A | 5141 | 0.19 | trans |
Section II. Transcriptome annotation
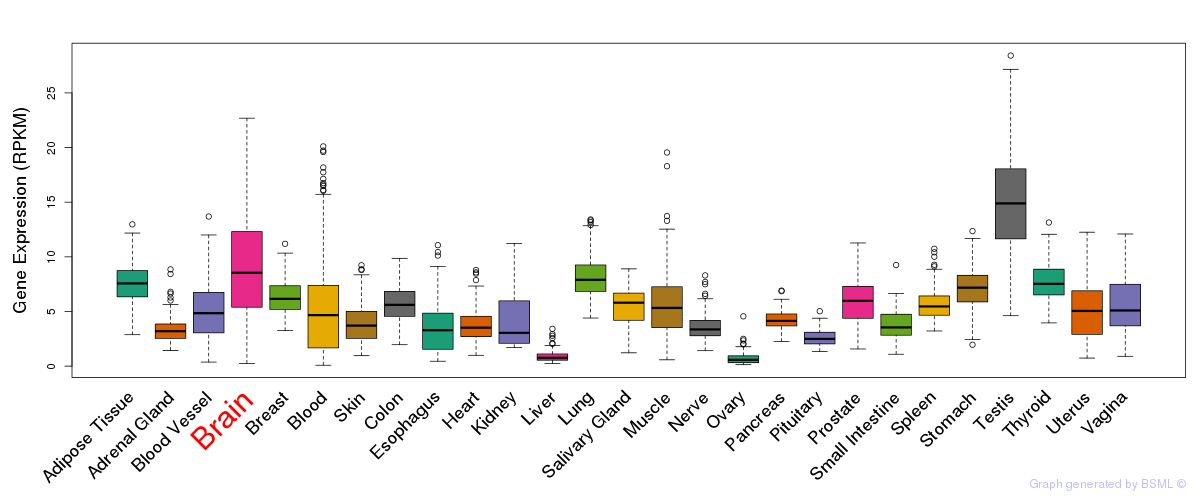
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC134043.1 | 0.93 | 0.12 |
| EBF1 | 0.93 | 0.28 |
| RARB | 0.91 | 0.62 |
| PBX3 | 0.89 | 0.35 |
| IKZF1 | 0.84 | 0.37 |
| GPR149 | 0.84 | 0.58 |
| C10orf41 | 0.81 | 0.59 |
| DLX6 | 0.78 | 0.55 |
| ZNF521 | 0.75 | 0.51 |
| DACH1 | 0.75 | 0.50 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDADC1 | -0.23 | 0.23 |
| EMID1 | -0.18 | -0.20 |
| CCDC90A | -0.18 | -0.15 |
| GTDC1 | -0.17 | 0.16 |
| IGSF21 | -0.17 | 0.01 |
| RHCG | -0.17 | -0.02 |
| GARNL3 | -0.17 | 0.05 |
| CHPT1 | -0.17 | 0.09 |
| SLC26A4 | -0.17 | 0.20 |
| NXPH3 | -0.16 | -0.05 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity | NAS | 8413254 | |
| GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity | TAS | 9677330 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 8940140 | |
| GO:0009187 | cyclic nucleotide metabolic process | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005624 | membrane fraction | TAS | 9677330 | |
| GO:0005625 | soluble fraction | TAS | 8940140 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 17 | 25 | 12 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1368 | 1375 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 907 | 914 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 907 | 913 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-135 | 1245 | 1251 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-137 | 1083 | 1090 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| miR-139 | 314 | 321 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| hsa-miR-139brain | UCUACAGUGCACGUGUCU | ||||
| miR-144 | 1369 | 1375 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-188 | 230 | 236 | m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-7 | 612 | 618 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.