Gene Page: TRIM33
Summary ?
| GeneID | 51592 |
| Symbol | TRIM33 |
| Synonyms | ECTO|PTC7|RFG7|TF1G|TIF1G|TIF1GAMMA|TIFGAMMA |
| Description | tripartite motif containing 33 |
| Reference | MIM:605769|HGNC:HGNC:16290|Ensembl:ENSG00000197323|HPRD:10423|Vega:OTTHUMG00000011891 |
| Gene type | protein-coding |
| Map location | 1p13.1 |
| Pascal p-value | 0.014 |
| Sherlock p-value | 0.757 |
| Fetal beta | 0.738 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TRIM33 | chr1 | 114967255 | G | A | NM_015906 NM_033020 | . . | silent silent | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
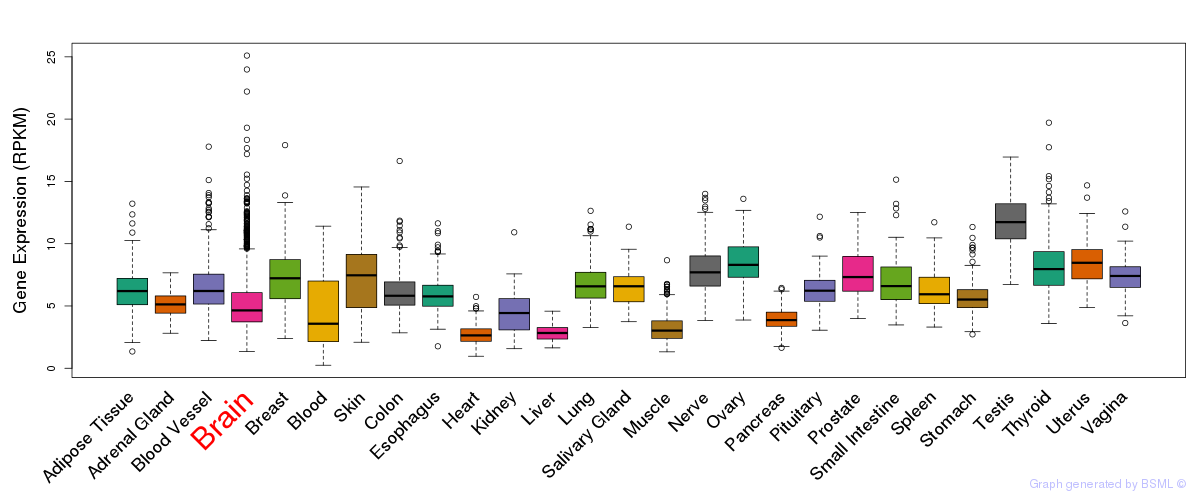
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | NAS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0016481 | negative regulation of transcription | NAS | 10022127 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | NAS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCERS KINOME GRAY | 15 | 10 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| FERRARI RESPONSE TO FENRETINIDE UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS DN | 53 | 29 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 3190 | 3196 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-137 | 3669 | 3676 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-19 | 4543 | 4550 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 145 | 152 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 3942 | 3949 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-21 | 1032 | 1038 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-214 | 3565 | 3571 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-24 | 1018 | 1024 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-26 | 1322 | 1328 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-5p | 3932 | 3938 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 1037 | 1043 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-338 | 3798 | 3805 | 1A,m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-370 | 3195 | 3201 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-384 | 3738 | 3744 | 1A | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-409-3p | 1107 | 1113 | 1A | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-455 | 4546 | 4552 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-491 | 1317 | 1323 | m8 | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.