Gene Page: CYB5R2
Summary ?
| GeneID | 51700 |
| Symbol | CYB5R2 |
| Synonyms | B5R.2 |
| Description | cytochrome b5 reductase 2 |
| Reference | MIM:608342|HGNC:HGNC:24376|Ensembl:ENSG00000166394|HPRD:06435|Vega:OTTHUMG00000165665 |
| Gene type | protein-coding |
| Map location | 11p15.4 |
| Pascal p-value | 0.59 |
| Sherlock p-value | 0.024 |
| Fetal beta | -2.142 |
| eGene | Cerebellar Hemisphere Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7580088 | chr2 | 171733349 | CYB5R2 | 51700 | 0.06 | trans | ||
| rs1894921 | chr2 | 171737414 | CYB5R2 | 51700 | 0.06 | trans | ||
| rs4893866 | chr2 | 177363635 | CYB5R2 | 51700 | 0.05 | trans | ||
| rs973234 | chr3 | 189840350 | CYB5R2 | 51700 | 0.15 | trans | ||
| rs9307467 | chr4 | 120029331 | CYB5R2 | 51700 | 0.04 | trans | ||
| rs17152736 | chr7 | 79290748 | CYB5R2 | 51700 | 0.12 | trans | ||
| rs6482363 | chr10 | 24368607 | CYB5R2 | 51700 | 0.06 | trans | ||
| rs1341055 | chr10 | 112607604 | CYB5R2 | 51700 | 0.06 | trans | ||
| rs17109739 | chr14 | 80148280 | CYB5R2 | 51700 | 0.1 | trans | ||
| rs17109778 | chr14 | 80170572 | CYB5R2 | 51700 | 0.14 | trans | ||
| rs2072968 | chr20 | 39999915 | CYB5R2 | 51700 | 0.18 | trans | ||
| rs7276414 | chr21 | 16489575 | CYB5R2 | 51700 | 0.13 | trans | ||
| rs7888140 | chrX | 19825933 | CYB5R2 | 51700 | 0.07 | trans | ||
| rs4076909 | chrX | 71329672 | CYB5R2 | 51700 | 0.03 | trans | ||
| rs1857642 | chrX | 136154060 | CYB5R2 | 51700 | 0.07 | trans |
Section II. Transcriptome annotation
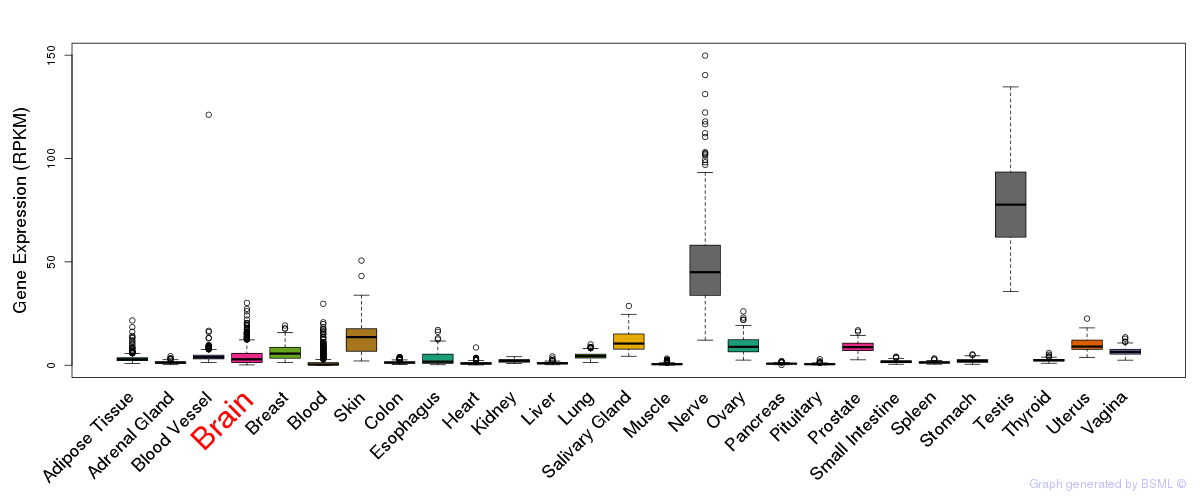
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN | 59 | 32 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |