Gene Page: CAB39
Summary ?
| GeneID | 51719 |
| Symbol | CAB39 |
| Synonyms | CGI-66|MO25 |
| Description | calcium binding protein 39 |
| Reference | MIM:612174|HGNC:HGNC:20292|Ensembl:ENSG00000135932|HPRD:10091|Vega:OTTHUMG00000133220 |
| Gene type | protein-coding |
| Map location | 2q37.1 |
| Pascal p-value | 0.84 |
| Sherlock p-value | 0.333 |
| Fetal beta | -0.469 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13313294 | 2 | 231646024 | CAB39 | 1.799E-4 | -0.334 | 0.034 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
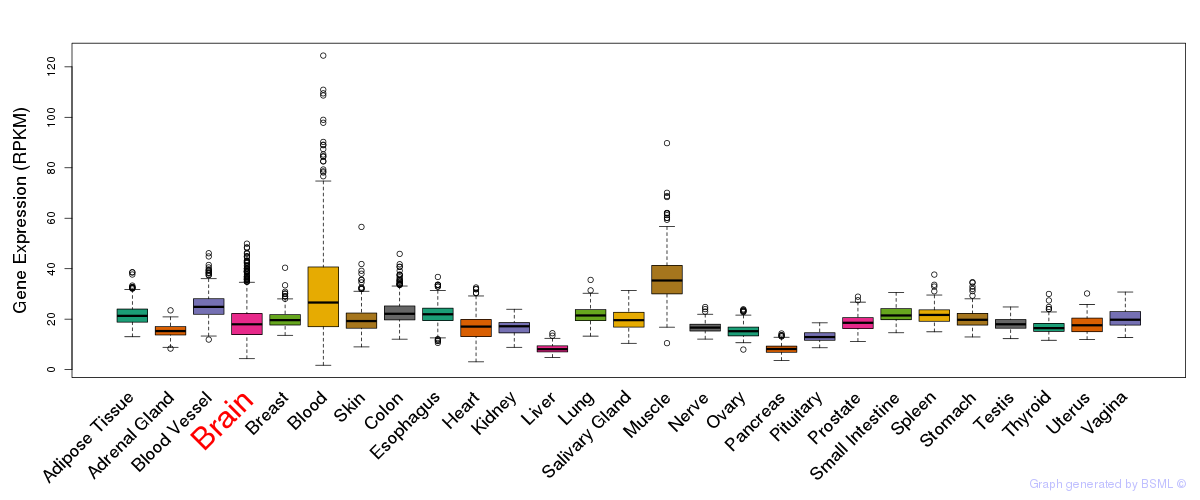
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCKDK | - | branched chain ketoacid dehydrogenase kinase | Affinity Capture-MS | BioGRID | 17353931 |
| CUL2 | MGC131970 | cullin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| FLJ10213 | - | hypothetical protein FLJ10213 | Affinity Capture-MS | BioGRID | 17353931 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| PRKAA1 | AMPK | AMPKa1 | MGC33776 | MGC57364 | protein kinase, AMP-activated, alpha 1 catalytic subunit | - | HPRD | 8910387 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | Affinity Capture-Western | BioGRID | 15561763 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | - | HPRD | 14676191 |
| STRADA | FLJ90524 | LYK5 | NY-BR-96 | PMSE | STRAD | Stlk | STE20-related kinase adaptor alpha | - | HPRD,BioGRID | 14730349 |15561763 |
| STRADB | ALS2CR2 | CALS-21 | ILPIP | ILPIPA | MGC102916 | PAPK | PRO1038 | STE20-related kinase adaptor beta | - | HPRD | 14676191 |
| TIMM13 | TIM13 | TIM13B | TIMM13A | TIMM13B | ppv1 | translocase of inner mitochondrial membrane 13 homolog (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | 19 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| ABDELMOHSEN ELAVL4 TARGETS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |