Gene Page: WWOX
Summary ?
| GeneID | 51741 |
| Symbol | WWOX |
| Synonyms | D16S432E|EIEE28|FOR|FRA16D|HHCMA56|PRO0128|SCAR12|SDR41C1|WOX1 |
| Description | WW domain containing oxidoreductase |
| Reference | MIM:605131|HGNC:HGNC:12799|Ensembl:ENSG00000186153|HPRD:05501|Vega:OTTHUMG00000176851 |
| Gene type | protein-coding |
| Map location | 16q23 |
| Fetal beta | -1.154 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27230472 | 16 | 78601767 | WWOX | 6.64E-5 | 0.549 | 0.024 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2707613 | 16 | 78048549 | WWOX | ENSG00000186153.12 | 7.68207E-7 | 0.03 | -84761 | gtex_brain_putamen_basal |
| rs2707612 | 16 | 78048960 | WWOX | ENSG00000186153.12 | 7.11006E-7 | 0.03 | -84350 | gtex_brain_putamen_basal |
| rs2735406 | 16 | 78049222 | WWOX | ENSG00000186153.12 | 5.02104E-7 | 0.03 | -84088 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
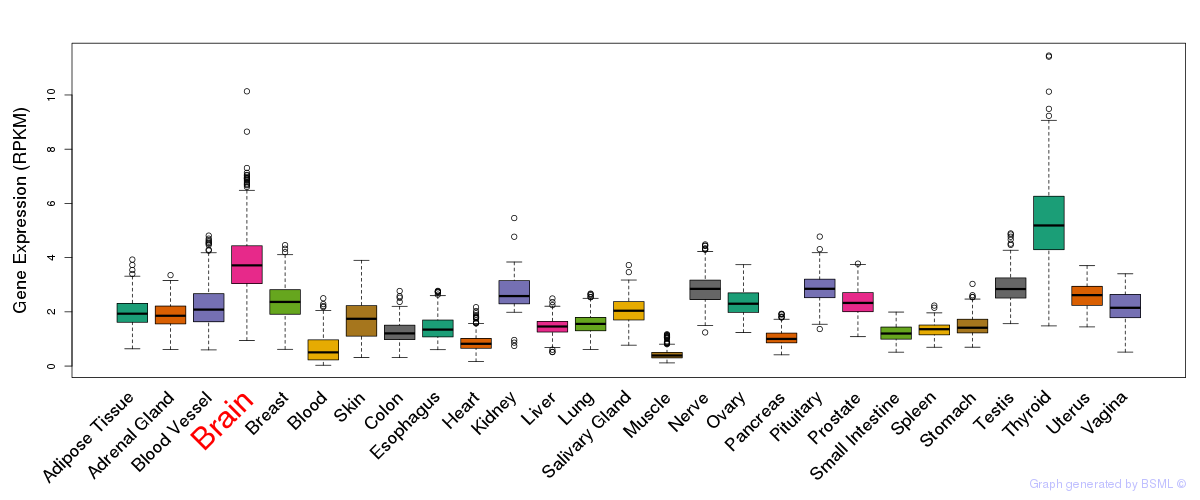
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| TAKADA GASTRIC CANCER COPY NUMBER DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES UP | 15 | 9 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION DN | 12 | 10 | All SZGR 2.0 genes in this pathway |