Gene Page: ERAP1
Summary ?
| GeneID | 51752 |
| Symbol | ERAP1 |
| Synonyms | A-LAP|ALAP|APPILS|ARTS-1|ARTS1|ERAAP|ERAAP1|PILS-AP|PILSAP |
| Description | endoplasmic reticulum aminopeptidase 1 |
| Reference | MIM:606832|HGNC:HGNC:18173|Ensembl:ENSG00000164307|HPRD:06015|Vega:OTTHUMG00000128721 |
| Gene type | protein-coding |
| Map location | 5q15 |
| Pascal p-value | 0.613 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09882587 | 5 | 96143769 | ERAP1 | 1.22E-9 | -0.026 | 1.3E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17482078 | 5 | 96118866 | ERAP1 | ENSG00000164307.8 | 2.518E-6 | 0.02 | 24937 | gtex_brain_putamen_basal |
| rs2124921 | 5 | 96119339 | ERAP1 | ENSG00000164307.8 | 2.518E-6 | 0.02 | 24464 | gtex_brain_putamen_basal |
| rs469758 | 5 | 96121715 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 22088 | gtex_brain_putamen_basal |
| rs30378 | 5 | 96121994 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 21809 | gtex_brain_putamen_basal |
| rs30379 | 5 | 96122260 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 21543 | gtex_brain_putamen_basal |
| rs30380 | 5 | 96122281 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 21522 | gtex_brain_putamen_basal |
| rs469367 | 5 | 96123055 | ERAP1 | ENSG00000164307.8 | 1.551E-6 | 0.02 | 20748 | gtex_brain_putamen_basal |
| rs168674 | 5 | 96123125 | ERAP1 | ENSG00000164307.8 | 4.113E-6 | 0.02 | 20678 | gtex_brain_putamen_basal |
| rs467735 | 5 | 96123160 | ERAP1 | ENSG00000164307.8 | 3.934E-6 | 0.02 | 20643 | gtex_brain_putamen_basal |
| rs246455 | 5 | 96123210 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 20593 | gtex_brain_putamen_basal |
| rs246454 | 5 | 96123264 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 20539 | gtex_brain_putamen_basal |
| rs246453 | 5 | 96123336 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 20467 | gtex_brain_putamen_basal |
| rs151964 | 5 | 96123666 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 20137 | gtex_brain_putamen_basal |
| rs30187 | 5 | 96124330 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 19473 | gtex_brain_putamen_basal |
| rs30186 | 5 | 96124447 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 19356 | gtex_brain_putamen_basal |
| rs28333 | 5 | 96124959 | ERAP1 | ENSG00000164307.8 | 3.99E-6 | 0.02 | 18844 | gtex_brain_putamen_basal |
| rs28337 | 5 | 96125314 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 18489 | gtex_brain_putamen_basal |
| rs30185 | 5 | 96125465 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 18338 | gtex_brain_putamen_basal |
| rs26510 | 5 | 96125910 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 17893 | gtex_brain_putamen_basal |
| rs27710 | 5 | 96126197 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 17606 | gtex_brain_putamen_basal |
| rs27529 | 5 | 96126308 | ERAP1 | ENSG00000164307.8 | 2.774E-6 | 0.02 | 17495 | gtex_brain_putamen_basal |
| rs27640 | 5 | 96127905 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 15898 | gtex_brain_putamen_basal |
| rs26512 | 5 | 96128400 | ERAP1 | ENSG00000164307.8 | 2.27E-6 | 0.02 | 15403 | gtex_brain_putamen_basal |
| rs27434 | 5 | 96129512 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 14291 | gtex_brain_putamen_basal |
| rs28119 | 5 | 96131733 | ERAP1 | ENSG00000164307.8 | 2.04E-6 | 0.02 | 12070 | gtex_brain_putamen_basal |
| rs26489 | 5 | 96133427 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 10376 | gtex_brain_putamen_basal |
| rs26500 | 5 | 96133712 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 10091 | gtex_brain_putamen_basal |
| rs26490 | 5 | 96133920 | ERAP1 | ENSG00000164307.8 | 8.316E-7 | 0.02 | 9883 | gtex_brain_putamen_basal |
| rs26491 | 5 | 96134113 | ERAP1 | ENSG00000164307.8 | 2.156E-6 | 0.02 | 9690 | gtex_brain_putamen_basal |
| rs149189 | 5 | 96137804 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 5999 | gtex_brain_putamen_basal |
| rs26495 | 5 | 96137840 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 5963 | gtex_brain_putamen_basal |
| rs26496 | 5 | 96137882 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 5921 | gtex_brain_putamen_basal |
| rs26497 | 5 | 96138767 | ERAP1 | ENSG00000164307.8 | 2.04E-6 | 0.02 | 5036 | gtex_brain_putamen_basal |
| rs26498 | 5 | 96138812 | ERAP1 | ENSG00000164307.8 | 2.166E-6 | 0.02 | 4991 | gtex_brain_putamen_basal |
| rs27045 | 5 | 96138861 | ERAP1 | ENSG00000164307.8 | 3.943E-6 | 0.02 | 4942 | gtex_brain_putamen_basal |
| rs151823 | 5 | 96159992 | ERAP1 | ENSG00000164307.8 | 2.176E-6 | 0.02 | -16189 | gtex_brain_putamen_basal |
| rs12519842 | 5 | 96194255 | ERAP1 | ENSG00000164307.8 | 1.532E-6 | 0.02 | -50452 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
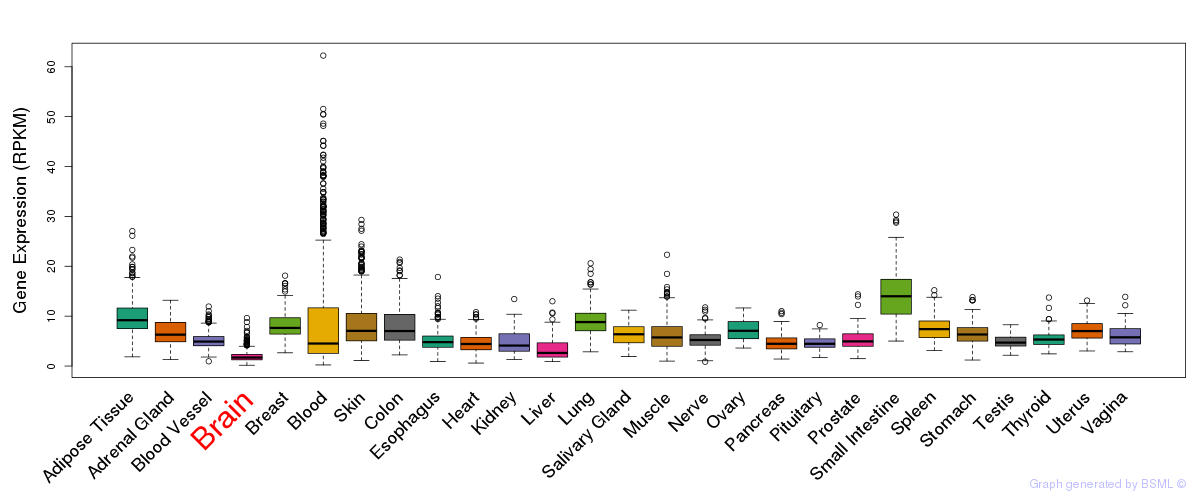
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION DN | 49 | 38 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| NICK RESPONSE TO PROC TREATMENT DN | 27 | 18 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |