Gene Page: PLA2G1B
Summary ?
| GeneID | 5319 |
| Symbol | PLA2G1B |
| Synonyms | PLA2|PLA2A|PPLA2 |
| Description | phospholipase A2 group IB |
| Reference | MIM:172410|HGNC:HGNC:9030|Ensembl:ENSG00000170890|HPRD:01396|Vega:OTTHUMG00000169343 |
| Gene type | protein-coding |
| Map location | 12q24.31 |
| Pascal p-value | 0.3 |
| Fetal beta | -0.949 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
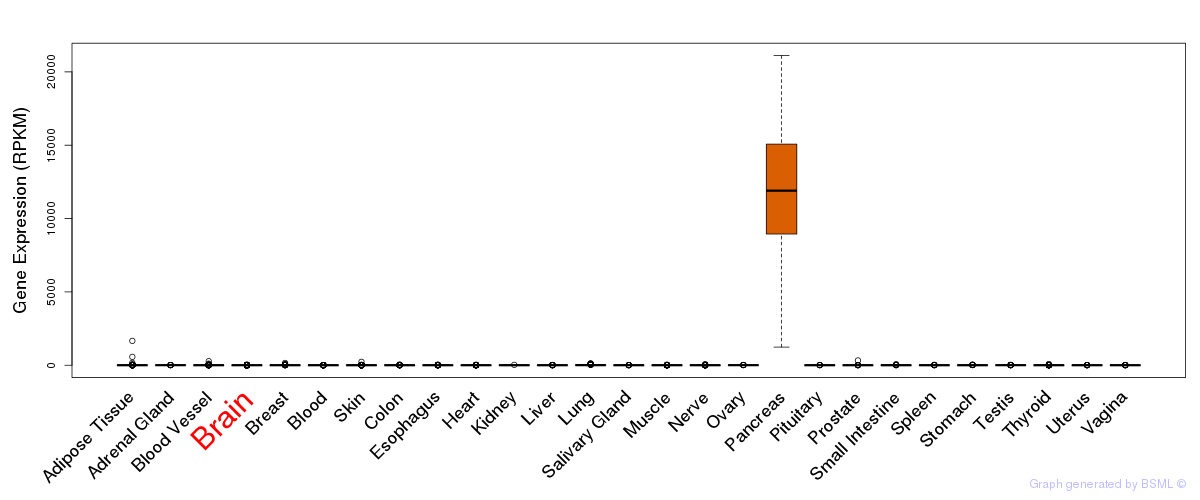
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRN | 0.78 | 0.80 |
| TRIM8 | 0.78 | 0.76 |
| LARS2 | 0.75 | 0.70 |
| NCSTN | 0.73 | 0.67 |
| KIAA2013 | 0.72 | 0.71 |
| SHC1 | 0.72 | 0.70 |
| SIDT2 | 0.72 | 0.72 |
| C2orf18 | 0.72 | 0.72 |
| SLC35A4 | 0.72 | 0.71 |
| ATG2A | 0.72 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYCP3 | -0.54 | -0.51 |
| AL050337.1 | -0.51 | -0.44 |
| FAM159B | -0.50 | -0.56 |
| AF347015.21 | -0.48 | -0.38 |
| AC120053.1 | -0.45 | -0.36 |
| SPDYA | -0.44 | -0.37 |
| RP9P | -0.44 | -0.43 |
| NOX1 | -0.44 | -0.35 |
| GNG11 | -0.43 | -0.31 |
| ACSM3 | -0.43 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IDA | Neurotransmitter (GO term level: 4) | 1918029 |
| GO:0004623 | phospholipase A2 activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0032052 | bile acid binding | ISS | 7060561 | |
| GO:0043498 | cell surface binding | IDA | 1918029 | |
| GO:0047498 | calcium-dependent phospholipase A2 activity | IDA | 7060561 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000187 | activation of MAPK activity | ISS | 15528384 |16005851 | |
| GO:0002446 | neutrophil mediated immunity | ISS | 16005851 | |
| GO:0016042 | lipid catabolic process | IEA | - | |
| GO:0007243 | protein kinase cascade | ISS | 15528384 | |
| GO:0010552 | positive regulation of specific transcription from RNA polymerase II promoter | ISS | 15528384 | |
| GO:0010524 | positive regulation of calcium ion transport into cytosol | ISS | 15528384 | |
| GO:0007165 | signal transduction | TAS | 9886417 | |
| GO:0015758 | glucose transport | ISS | 1918029 | |
| GO:0007015 | actin filament organization | TAS | 9886417 | |
| GO:0006644 | phospholipid metabolic process | IEA | - | |
| GO:0006633 | fatty acid biosynthetic process | IDA | 1918029 | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | ISS | 15528384 | |
| GO:0032637 | interleukin-8 production | ISS | 15528384 | |
| GO:0032869 | cellular response to insulin stimulus | ISS | 1918029 | |
| GO:0019370 | leukotriene biosynthetic process | ISS | 16005851 | |
| GO:0030593 | neutrophil chemotaxis | ISS | 16005851 | |
| GO:0032431 | activation of phospholipase A2 | TAS | 17981679 | |
| GO:0045740 | positive regulation of DNA replication | IDA | 1918029 | |
| GO:0048146 | positive regulation of fibroblast proliferation | IC | 1918029 | |
| GO:0050714 | positive regulation of protein secretion | TAS | 17981679 | |
| GO:0050778 | positive regulation of immune response | ISS | 16005851 | |
| GO:0044240 | multicellular organismal lipid catabolic process | IDA | 7060561 | |
| GO:0046470 | phosphatidylcholine metabolic process | IDA | 7060561 | |
| GO:0050482 | arachidonic acid secretion | TAS | 12423354 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 7060561 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCEROPHOSPHOLIPID METABOLISM | 77 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG ETHER LIPID METABOLISM | 33 | 15 | All SZGR 2.0 genes in this pathway |
| KEGG ARACHIDONIC ACID METABOLISM | 58 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LINOLEIC ACID METABOLISM | 29 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG ALPHA LINOLENIC ACID METABOLISM | 19 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PI | 15 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PC | 22 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PA | 27 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PG | 16 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PE | 21 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PS | 15 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | 82 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |