Gene Page: PLD2
Summary ?
| GeneID | 5338 |
| Symbol | PLD2 |
| Synonyms | - |
| Description | phospholipase D2 |
| Reference | MIM:602384|HGNC:HGNC:9068|Ensembl:ENSG00000129219|HPRD:03857|Vega:OTTHUMG00000090779 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.207 |
| Sherlock p-value | 0.268 |
| Fetal beta | 0.229 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23956071 | 17 | 4710044 | PLD2 | 1.131E-4 | -0.367 | 0.029 | DMG:Wockner_2014 |
| cg16685860 | 17 | 4710619 | PLD2 | -0.03 | 0.46 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1038121 | chr17 | 4475243 | PLD2 | 5338 | 0.12 | cis | ||
| rs1038118 | chr17 | 4475583 | PLD2 | 5338 | 0.14 | cis | ||
| rs16905508 | chr8 | 137113905 | PLD2 | 5338 | 0.08 | trans | ||
| rs11785077 | chr8 | 137115681 | PLD2 | 5338 | 0.08 | trans |
Section II. Transcriptome annotation
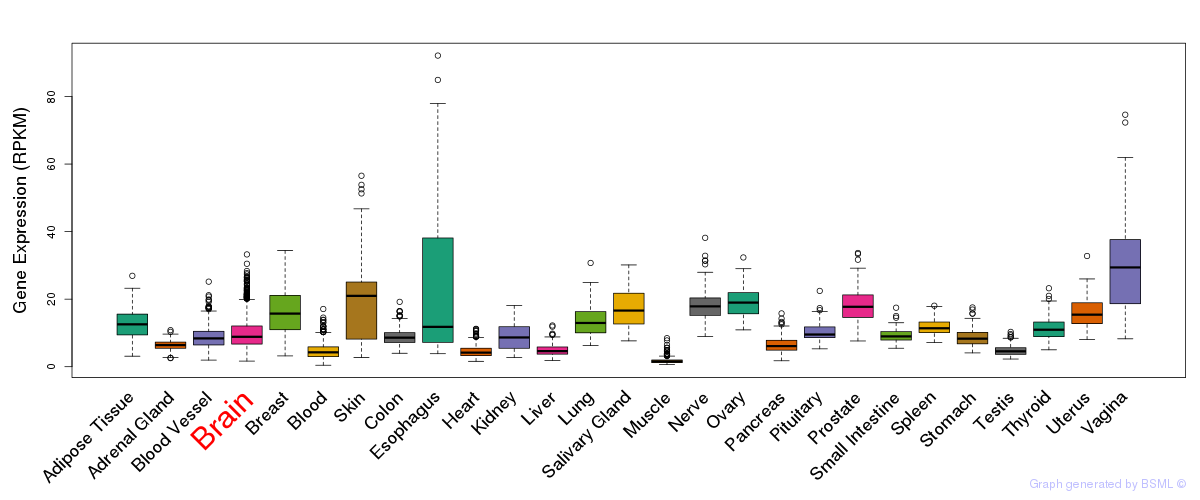
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 11373276 |
| ALDOA | ALDA | MGC10942 | MGC17716 | MGC17767 | aldolase A, fructose-bisphosphate | - | HPRD,BioGRID | 11876650 |
| AMPH | AMPH1 | amphiphysin | - | HPRD,BioGRID | 10764771 |
| AQP3 | GIL | aquaporin 3 (Gill blood group) | - | HPRD,BioGRID | 14675200 |
| ARF1 | - | ADP-ribosylation factor 1 | Reconstituted Complex | BioGRID | 10801846 |11373276 |
| ARF1 | - | ADP-ribosylation factor 1 | - | HPRD | 10801846 |12519790 |
| BIN1 | AMPH2 | AMPHL | DKFZp547F068 | MGC10367 | SH3P9 | bridging integrator 1 | - | HPRD,BioGRID | 10764771 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 10675563 |
| CAV2 | CAV | MGC12294 | caveolin 2 | Co-fractionation | BioGRID | 10675563 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD | 12697812 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9837959 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with PLD1. This interaction was modelled on a demonstrated interaction between human EGFR and mouse PLD2. | BIND | 9837959 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 16837165 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD | 12697812 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | - | HPRD,BioGRID | 12753082 |
| GM2A | SAP-3 | GM2 ganglioside activator | - | HPRD | 11672434 |
| KIAA1303 | - | raptor | Affinity Capture-Western | BioGRID | 16837165 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 12697812 |
| OPRM1 | KIAA0403 | MOR | MOR1 | OPRM | opioid receptor, mu 1 | - | HPRD,BioGRID | 12519790 |
| PEA15 | HMAT1 | HUMMAT1H | MAT1 | MAT1H | PEA-15 | PED | phosphoprotein enriched in astrocytes 15 | - | HPRD,BioGRID | 10926929 |
| PIP5K1A | - | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | - | HPRD | 11032811 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 12646582 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | PLC-gamma1 interacts with PLD2. This interaction was modeled on a demonstrated interaction between PLC-gamma1 from an unspecified species and human PLD2. | BIND | 14744865 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 11042115 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | - | HPRD,BioGRID | 11744693 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 12697812 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCEROPHOSPHOLIPID METABOLISM | 77 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG ETHER LIPID METABOLISM | 33 | 15 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST GAQ PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID ARF 3PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PA | 27 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | 82 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 17P13 DELETION | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |