Gene Page: CLIC6
Summary ?
| GeneID | 54102 |
| Symbol | CLIC6 |
| Synonyms | CLIC1L |
| Description | chloride intracellular channel 6 |
| Reference | MIM:615321|HGNC:HGNC:2065|Ensembl:ENSG00000159212|Vega:OTTHUMG00000086237 |
| Gene type | protein-coding |
| Map location | 21q22.12 |
| Pascal p-value | 0.395 |
| Fetal beta | 0.31 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2573 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18074297 | 21 | 36041612 | CLIC6 | -0.02 | 0.55 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | CLIC6 | 54102 | 0 | trans | ||
| rs1736557 | chr1 | 171080079 | CLIC6 | 54102 | 0.15 | trans | ||
| rs2922180 | chr3 | 125280472 | CLIC6 | 54102 | 0.04 | trans | ||
| rs2976809 | chr3 | 125451738 | CLIC6 | 54102 | 0.02 | trans | ||
| rs6441080 | chr3 | 156298934 | CLIC6 | 54102 | 0.1 | trans | ||
| rs7615297 | chr3 | 156299312 | CLIC6 | 54102 | 0.19 | trans | ||
| rs687188 | chr4 | 130527157 | CLIC6 | 54102 | 0.09 | trans | ||
| rs639864 | chr4 | 130538020 | CLIC6 | 54102 | 0.05 | trans | ||
| rs12663450 | chr6 | 38135729 | CLIC6 | 54102 | 0.09 | trans | ||
| rs6455226 | chr6 | 67930295 | CLIC6 | 54102 | 0.18 | trans | ||
| rs2815732 | chr6 | 73151761 | CLIC6 | 54102 | 0.19 | trans | ||
| rs283063 | chr6 | 118621313 | CLIC6 | 54102 | 0.19 | trans | ||
| rs3823550 | chr7 | 137346101 | CLIC6 | 54102 | 0.01 | trans | ||
| rs10958899 | chr9 | 10128700 | CLIC6 | 54102 | 0.2 | trans | ||
| rs10958907 | chr9 | 10131580 | CLIC6 | 54102 | 0.11 | trans | ||
| rs10958910 | chr9 | 10134759 | CLIC6 | 54102 | 0.05 | trans | ||
| rs427497 | chr9 | 107982791 | CLIC6 | 54102 | 0.13 | trans | ||
| rs9949772 | chr18 | 31042632 | CLIC6 | 54102 | 0.05 | trans | ||
| rs11873703 | chr18 | 61448702 | CLIC6 | 54102 | 0.03 | trans | ||
| rs6095741 | chr20 | 48666589 | CLIC6 | 54102 | 7.292E-5 | trans | ||
| rs7062071 | chrX | 17724830 | CLIC6 | 54102 | 0.03 | trans | ||
| rs1545676 | chrX | 93530472 | CLIC6 | 54102 | 0.03 | trans | ||
| rs35990795 | 21 | 36085834 | CLIC6 | ENSG00000159212.8 | 3.295E-6 | 0.05 | 44146 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
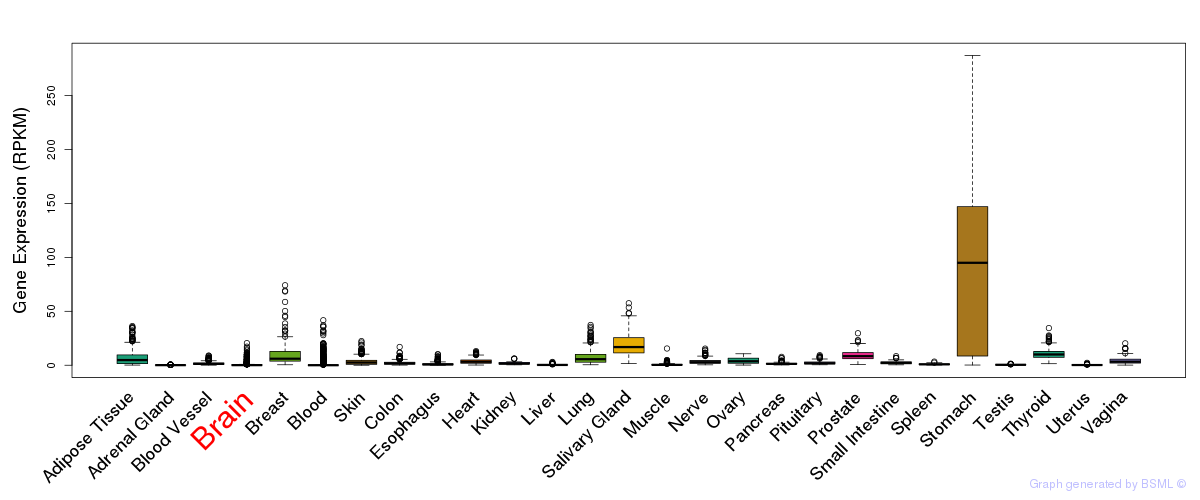
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005247 | voltage-gated chloride channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006821 | chloride transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA | 39 | 25 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 4 UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 1699 | 1705 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.