Gene Page: PNN
Summary ?
| GeneID | 5411 |
| Symbol | PNN |
| Synonyms | DRS|DRSP|SDK3|memA |
| Description | pinin, desmosome associated protein |
| Reference | MIM:603154|HGNC:HGNC:9162|Ensembl:ENSG00000100941|HPRD:04401|Vega:OTTHUMG00000028821 |
| Gene type | protein-coding |
| Map location | 14q21.1 |
| Pascal p-value | 0.511 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.09:CC_BA10_disease_P=0.0011:HBB_BA9_fold_change=1.19:HBB_BA9_disease_P=0.0151 |
| Fetal beta | 1.263 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
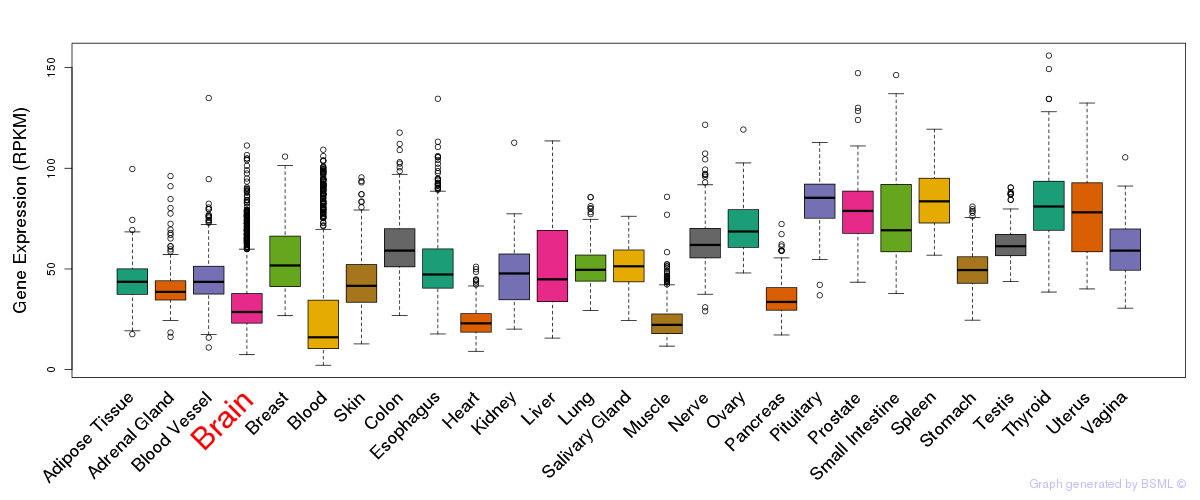
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LSMD1 | 0.77 | 0.56 |
| BRP44L | 0.73 | 0.49 |
| PSMB10 | 0.73 | 0.58 |
| CNFN | 0.72 | 0.45 |
| NDUFB7 | 0.71 | 0.44 |
| MYEOV2 | 0.70 | 0.48 |
| A1BG | 0.69 | 0.23 |
| NDUFA1 | 0.69 | 0.37 |
| NDUFA13 | 0.69 | 0.48 |
| CHCHD10 | 0.68 | 0.48 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF418 | -0.40 | -0.54 |
| ZNF211 | -0.38 | -0.50 |
| PTPN12 | -0.38 | -0.48 |
| ZNF841 | -0.37 | -0.51 |
| ZNF589 | -0.37 | -0.50 |
| THOC2 | -0.36 | -0.46 |
| ERCC5 | -0.36 | -0.45 |
| USP34 | -0.36 | -0.43 |
| ZNF17 | -0.36 | -0.49 |
| MLL5 | -0.36 | -0.49 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | - | HPRD,BioGRID | 15542832 |
| CTBP2 | - | C-terminal binding protein 2 | - | HPRD | 15542832 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD,BioGRID | 10809736 |
| KRT19 | CK19 | K19 | K1CS | MGC15366 | keratin 19 | - | HPRD,BioGRID | 10809736 |
| KRT8 | CARD2 | CK8 | CYK8 | K2C8 | K8 | KO | keratin 8 | - | HPRD,BioGRID | 10809736 |
| NFKBIL1 | IKBL | LST1 | NFKBIL | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PPIG | CARS-Cyp | CYP | MGC133241 | SRCyp | peptidylprolyl isomerase G (cyclophilin G) | - | HPRD,BioGRID | 15358154 |
| PRPF4B | KIAA0536 | PR4H | PRP4 | PRP4H | PRP4K | dJ1013A10.1 | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | - | HPRD,BioGRID | 12077342 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | - | HPRD,BioGRID | 14517304 |
| SFRS18 | C6orf111 | DKFZp564B0769 | FLJ14752 | FLJ14853 | FLJ14992 | FLJ90147 | HSPC306 | MGC104269 | RP11-98I9.2 | SRrp130 | bA98I9.2 | splicing factor, arginine/serine-rich 18 | Pnn interacts with SRrp130. | BIND | 14578391 |
| SFRS18 | C6orf111 | DKFZp564B0769 | FLJ14752 | FLJ14853 | FLJ14992 | FLJ90147 | HSPC306 | MGC104269 | RP11-98I9.2 | SRrp130 | bA98I9.2 | splicing factor, arginine/serine-rich 18 | - | HPRD,BioGRID | 14578391 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | - | HPRD,BioGRID | 14578391 |
| SFRS4 | SRP75 | splicing factor, arginine/serine-rich 4 | Pnn interacts with SRp75. | BIND | 14578391 |
| SRRM2 | 300-KD | CWF21 | DKFZp686O15166 | FLJ21926 | FLJ22250 | KIAA0324 | MGC40295 | SRL300 | SRm300 | serine/arginine repetitive matrix 2 | Pnn interacts with SRm300. | BIND | 14578391 |
| SRRM2 | 300-KD | CWF21 | DKFZp686O15166 | FLJ21926 | FLJ22250 | KIAA0324 | MGC40295 | SRL300 | SRm300 | serine/arginine repetitive matrix 2 | - | HPRD,BioGRID | 14578391 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
| ZCCHC17 | HSPC251 | PS1D | RP11-266K22.1 | pNO40 | zinc finger, CCHC domain containing 17 | - | HPRD,BioGRID | 12893261 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER WITH BRCA1 MUTATED UP | 56 | 27 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| YUAN ZNF143 PARTNERS | 22 | 15 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL UP | 38 | 22 | All SZGR 2.0 genes in this pathway |