Gene Page: SEPT5
Summary ?
| GeneID | 5413 |
| Symbol | SEPT5 |
| Synonyms | CDCREL|CDCREL-1|CDCREL1|H5|HCDCREL-1|PNUTL1 |
| Description | septin 5 |
| Reference | MIM:602724|HGNC:HGNC:9164|Ensembl:ENSG00000184702|HPRD:04100|Vega:OTTHUMG00000150399 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Sherlock p-value | 0.334 |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.human_TAP-PSD-95-CORE CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3501 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
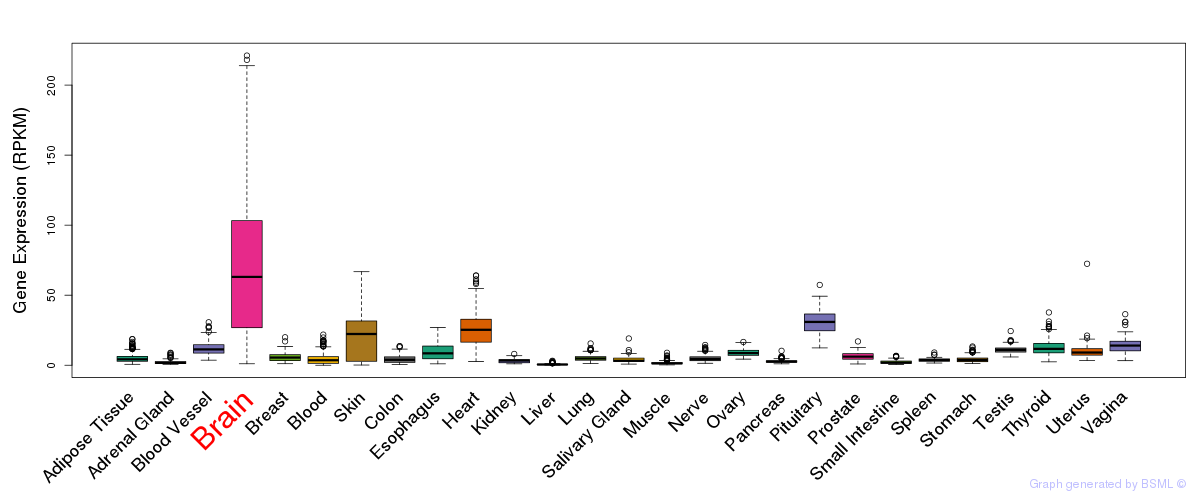
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HRSP12 | 0.89 | 0.89 |
| BBOX1 | 0.88 | 0.93 |
| SLC15A2 | 0.86 | 0.87 |
| DBX2 | 0.84 | 0.82 |
| SLCO1C1 | 0.83 | 0.90 |
| HSD17B6 | 0.83 | 0.91 |
| S1PR1 | 0.83 | 0.85 |
| GLUD1 | 0.82 | 0.78 |
| PLSCR4 | 0.82 | 0.76 |
| CD302 | 0.81 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM131B | -0.64 | -0.76 |
| KIF3C | -0.63 | -0.74 |
| NMNAT2 | -0.63 | -0.74 |
| NEURL4 | -0.63 | -0.75 |
| RTF1 | -0.63 | -0.74 |
| C6orf168 | -0.62 | -0.75 |
| RAB35 | -0.62 | -0.76 |
| MTMR4 | -0.62 | -0.75 |
| PREP | -0.62 | -0.73 |
| EIF4ENIF1 | -0.62 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 9385360 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10321247 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005198 | structural molecule activity | TAS | 9611266 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016080 | synaptic vesicle targeting | TAS | Synap (GO term level: 10) | 10321247 |
| GO:0000910 | cytokinesis | TAS | 9611266 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0017157 | regulation of exocytosis | IMP | 10321247 | |
| GO:0017157 | regulation of exocytosis | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IDA | Synap, Neurotransmitter (GO term level: 12) | 10321247 |
| GO:0008021 | synaptic vesicle | ISS | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0005886 | plasma membrane | IDA | 10321247 | |
| GO:0005886 | plasma membrane | ISS | - | |
| GO:0031105 | septin complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PARKIN PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 22 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-29 | 1538 | 1544 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.